Annotate with Exon Numbers
To run the Annotate with Exon Numbers tool, go to:
Toolbox | Utility Tools (![]() ) | Tracks (
) | Tracks (![]() ) | Annotate and Filter (
) | Annotate and Filter (![]() ) | Annotate with Exon Numbers (
) | Annotate with Exon Numbers (![]() )
)
Given a track with mRNA or CDS annotations, a new track will be created in which variants or other track items are annotated with the numbering of the corresponding exon with numbered exons based on the transcript annotations and gene names in the input track (see an example of a result in figure 25.23).
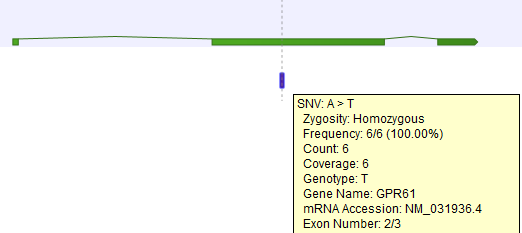
Figure 25.23: A variant found in the second exon out of three in total.
When there are multiple isoforms, a comma-separated list of the exon numbers is given. If an annotation overlaps an intron and has no partial overlap with an exon, this is indicated with a dash. Examples of exon annotations:
- [3/7] The annotation overlaps exon 3 in a gene that has 7 exons.
- [-/4] The annotation overlaps an intron in a gene that has 4 exons.
- [-/4, 38/38] The annotation overlaps an intron in a gene that has 4 exons as well as exon 38 in a gene with 38 exons.
- [12..17/29] The annotation overlaps exons 12 to 17 in a gene that has 29 exons.
- [12..9/18] The annotation overlaps exons 9 to 12 in a gene that has 18 exons. Exon 12 is written before exon 9, because the gene is located on the reverse strand.
The option One transcript per gene chooses a single transcript per gene. So if a variant overlaps multiple transcripts of the same gene, the tool chooses one of these transcripts and only shows overlapping exon numbers for one transcript per gene. The single transcript per gene that is used is chosen based on multiple criteria: First choose the transcript with best priority (lowest priority number). If none of the transcripts has a priority, or multiple transcripts have the same priority, then choose the transcript with the most exons overlapping the input item. Then choose the longest transcript, meaning the one with the highest sum of exon lengths. And finally choose the lexicographically first transcript id.
