Split Sequence List
Split Sequence List can split up nucleotide or peptide sequence lists. The output can be a specified number of lists, lists containing a specified number of sequences, or lists containing sequences with particular attribute values, such as terms in the description,
To launch the Split Sequence List tool, go to:
Toolbox | Utility Tools (![]() ) | Sequence Lists (
) | Sequence Lists (![]() ) |Split Sequence List (
) |Split Sequence List (![]() )
)
Select one or more sequence lists as input. If multiple lists are selected, then to process each input list separately, check the "Batch" checkbox. When unchecked, sequences in all the lists are considered together as a single input.
In the next wizard step, the basis upon which to do the splitting and any necessary settings for that option are specified (figure 35.14).
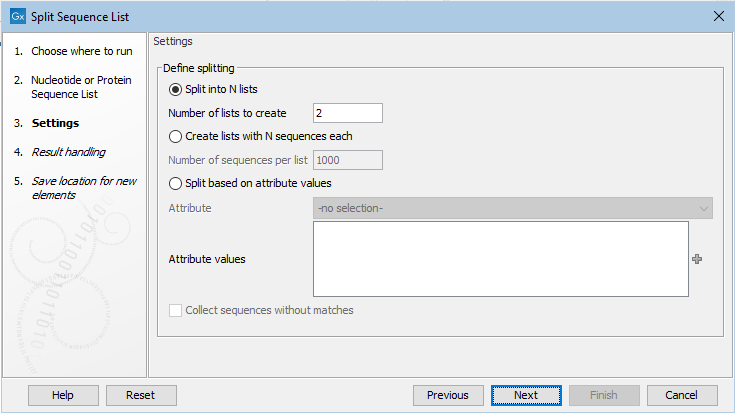
Figure 35.14: Sequence lists can be split into a set number of groups, or into lists containing particular numbers of sequences, or split based on attribute values.
The options are:
- Split into N lists In the "Number of lists to create" box, enter the number of lists to split the input into.
- Create lists with N sequences each In the "Number of sequences per list" box, enter the relevant number. The final sequence list in the set created may contain fewer than this number.
- Split based on attribute values Specify the attribute to split upon from the drop-down list. Columns in the table view of a sequence list equate to the attributes that the list can be split upon.
If no information is entered into the "Attribute values" field, a sequence list is created for each unique value of the specified attribute. If values are provided, a sequence list is created for each of these where at least one sequence has that attribute value. For example, if 3 values are specified, and sequences were found with attributes matching each of these values, 3 sequence lists would be created. If no sequences were found containing 1 of those attribute values, then only 2 sequence lists would be created. Check the "Collect sequences without matches" box to additionally produce a sequence list containing the sequences where no match to a specified value was identified.
Attribute values should be added in a new-line separated list (figure 35.15).
Matching of values is case specific. An asterisk should be added before and/or after the term if the value is part of a longer entry (figure 35.15).
Attribute values are matched in the order they are listed. For example, if the attribute values shown in figure 35.15 were used, and a sequence had a description with both terms in it, that sequence would be placed in the list containing sequences with "Putative" in their descriptions. This is because
*Putative*is listed first in the list of values provided to the tool.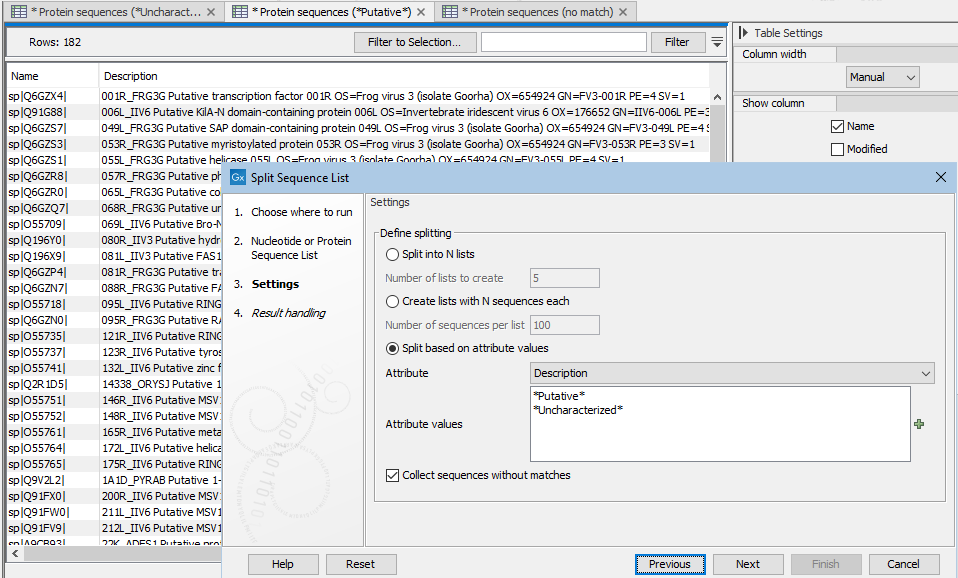
Figure 35.15: With the settings shown here, 3 sequence lists were created. These lists are open in the background tabs shown. One contains sequences with descriptions that include the term "Putative", one contains sequences with descriptions that include the term "Uncharacterized", and one contains sequences containing neither term in the desccription.
