GEO (Gene Expression Omnibus)
The GEO (Gene Expression Omnibus) sample and series formats are supported. Figure 37.10 shows how to download the data from GEO in the right format. GEO is located at http://www.ncbi.nlm.nih.gov/geo/.
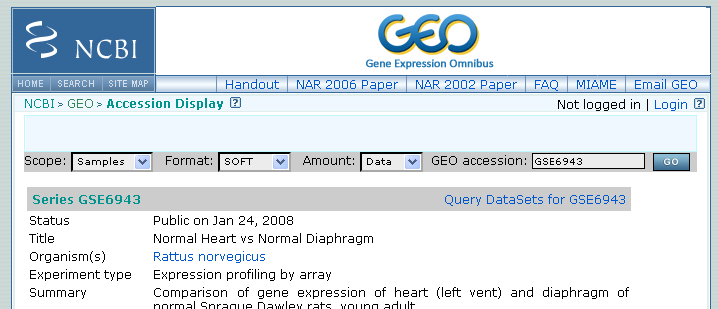
Figure 37.10: Selecting Samples, SOFT and Data before clicking go will give you the format supported by the CLC Genomics Workbench.
The GEO sample files are tab-delimited .txt files. They have three required lines:
^SAMPLE = GSM21610 !sample_table_begin ... !sample_table_endThe first line should start with
^SAMPLE = followed by the sample name, the line !sample_table_begin and the line !sample_table_end. Between the !sample_table_begin and !sample_table_end, lines are the column contents of the sample.
Note that GEO sample importer will also work for concatenated GEO sample files -- allowing multiple samples to be imported in one go. Download a sample file containing concatenated sample files here:
https://resources.qiagenbioinformatics.com/madata/GEOSampleFilesConcatenated.txt
Below you can find examples of the formatting of the GEO formats.
GEO sample file, simple
This format is very simple and includes two columns: one for feature id (e.g. gene name) and one for the expression value.
^SAMPLE = GSM21610 !sample_table_begin ID_REF VALUE id1 105.8 id2 32 id3 50.4 id4 57.8 id5 2914.1 !sample_table_endDownload the sample file here:
https://resources.qiagenbioinformatics.com/madata/GEOSampleFileSimple.txt
GEO sample file, including present/absent calls
This format includes an extra column for absent/present calls that can also be imported.
^SAMPLE = GSM21610 !sample_table_begin ID_REF VALUE ABS_CALL id1 105.8 M id2 32 A id3 50.4 A id4 57.8 A id5 2914.1 P !sample_table_endDownload the sample file here:
https://resources.qiagenbioinformatics.com/madata/GEOSampleFileAbsentPresent.txt
GEO sample file, including present/absent calls and p-values
This format includes two extra columns: one for absent/present calls and one for absent/present call p-values, that can also be imported.
^SAMPLE = GSM21610 !sample_table_begin ID_REF VALUE ABS_CALL DETECTION P-VALUE id1 105.8 M 0.00227496 id2 32 A 0.354441 id3 50.4 A 0.904352 id4 57.8 A 0.937071 id5 2914.1 P 6.02111e-05 !sample_table_endDownload the sample file here:
https://resources.qiagenbioinformatics.com/madata/GEOSampleFileAbsentPresentCallAndPValue.txt
GEO sample file: using absent/present call and p-value columns for sequence information
The workbench assumes that if there is a third column in the GEO sample file then it contains present/absent calls and that if there is a fourth column then it contains p-values for these calls. This means that the contents of the third column is assumed to be text and that of the fourth column a number. As long as these two basic requirements are met, the sample should be recognized and interpreted correctly.
You can thus use these two columns to carry additional information on your probes. The absent/present column can be used to carry additional information like e.g. sequence tags as shown below:
^SAMPLE = GSM21610 !sample_table_begin ID_REF VALUE ABS_CALL id1 105.8 AAA id2 32 AAC id3 50.4 ATA id4 57.8 ATT id5 2914.1 TTA !sample_table_endDownload the sample file here:
https://resources.qiagenbioinformatics.com/madata/GEOSampleFileSimpleSequenceTag.txt
Or, if you have multiple probes per sequence you could use the present/absent column to hold the sequence name and the p-value column to hold the interrogation position of your probes:
^SAMPLE = GSM21610 !sample_table_begin ID_REF VALUE ABS_CALL DETECTION P-VALUE probe1 755.07 seq1 1452 probe2 587.88 seq1 497 probe3 716.29 seq1 1447 probe4 1287.18 seq2 1899 !sample_table_end
Download the sample file here: https://resources.qiagenbioinformatics.com/madata/GEOSampleFileSimpleSequenceTagAndProbe.txt
GEO series file, simple
The series file includes expression values for multiple samples. Each of the samples in the file will be represented by its own element with the sample name. The first row lists the sample names.
!Series_title "Myb specificity determinants" !series_matrix_table_begin "ID_REF" "GSM21610" "GSM21611" "GSM21612" "id1" 2541 1781.8 1804.8 "id2" 11.3 621.5 50.2 "id3" 61.2 149.1 22 "id4" 55.3 328.8 97.2 "id5" 183.8 378.3 423.2 !series_matrix_table_end
Download the sample file here: https://resources.qiagenbioinformatics.com/madata/GEOSeriesFile.txt
