Quality filters
- Base quality filter: The base quality filter can be used to ignore the reads whose nucleotide at the potential variant position is of dubious quality. This is assessed by considering the quality of the nucleotides in the region around the nucleotide position. There are three parameters to determine the base quality filter:
- Neighborhood radius: This parameter determines the region size. For example if a neighborhood radius of five is used, a nucleotide will be evaluated based on the nucleotides that are 5 positions upstream and 5 positions downstream of the examined site, for a total of 11 nucleotides. Note that, near the end of the reads, eleven nucleotides will still be considered by offsetting the region relative to the nucleotide in question.
- Minimum central quality: Reads whose central base has a quality below the specified value will be ignored. This parameter does not apply to deletions since there is no 'central base' in these cases.
- Minimum neighborhood quality: Reads for which the minimum quality of the bases is below the specified value will be ignored.
Figure 29.10 gives an example of a variant called when the base quality filter is NOT applied, and not called when it is. When switching on the 'Show quality scores' option in the side panel of the reads it becomes visible that the reads that carry the potential 'G' variant tend to have poor quality. Note that the error in the example shown is a 'typical' Illumina error: the reference has a 'T' that is surrounded by stretches of 'G', the 'G' signals 'drowning' the signal of the 'T'. As all reads that have a base with quality less than 20 in this potential variant position are ignored when the 'Base quality filter' is turned on, no variant is called, most likely because it now does not meet the requirements of either the 'Minimum coverage', 'Minimum count' or 'Minimum frequency' filters.
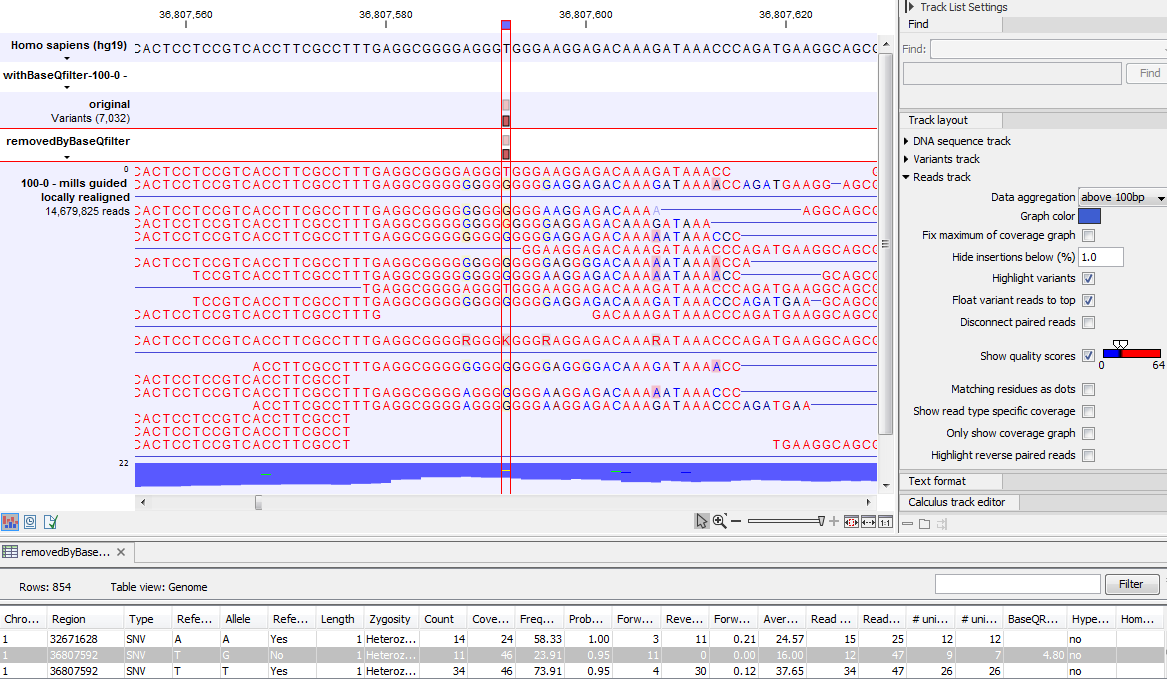
Figure 29.10: Example of a variant called when the base quality filter is NOT applied, and not called when it is.
