How a model structure is created
A structure model is created by mapping the query sequence onto the template structure based on a sequence alignment (see figure 18.19):
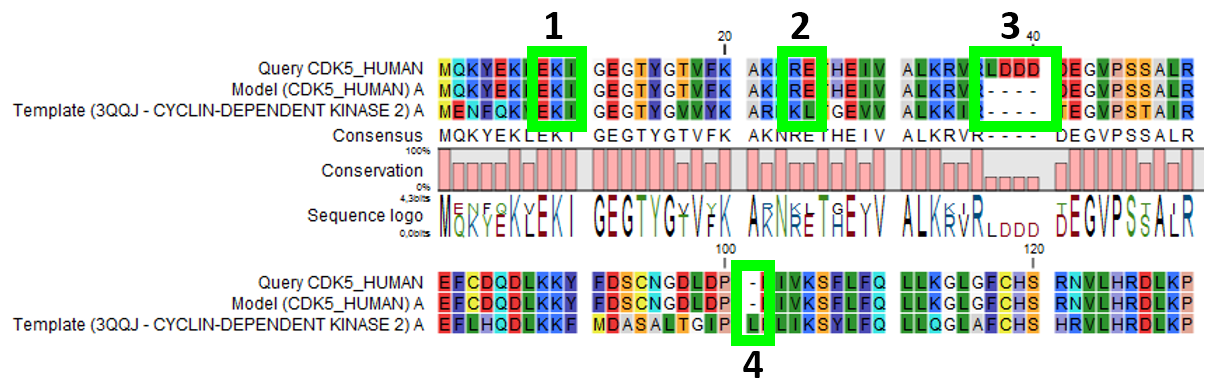
Figure 18.19: Sequence alignment mapping query sequence (Query CDK5_HUMAN) to the structure with sequence "Template(3QQJ - CYCLIN-DEPENDENT KINASE 2)", producing a structure with sequence "Model(CDK5_HUMAN)". Examples are highlighted: 1. Identical amino acids, 2. Amino acid changes, 3. Amino acids in query sequence not aligned to a position on the template structure, and 4. Amino acids on the template structure, not aligned to query sequence.
- For identical amino acids (example 1 in figure 18.19) => Copy atom positions from the PDB file. If the side chain is missing atoms in the PDB file, the side chain is rebuilt (How side chains are modeled).
- For amino acid changes (example 2 in figure 18.19) => Copy backbone atom positions from the PDB file. Model side chain atom positions to match the query sequence (How side chains are modeled).
- For amino acids in the query sequence not aligned to a position on the template structure (example 3 in figure 18.19) => No atoms are modeled. The model backbone will have a gap at this position and a "Structure modeling" issue is raised (see Import issues).
- For amino acids on the template structure, not aligned to the query sequence (example 4 in figure 18.19) => The residues are deleted from the structure and a "Structure modeling" issue is raised (see Import issues).
