Backbone
(For the molecules in the Proteins and Nucleic Acids categories, the backbone structure can be visualized in a schematic rendering, highlighting the secondary structure elements for proteins and matching base pairs for nucleic acids. The backbone visualization can be combined with any of the atom-level visualizations.
Five color schemes are available for backbone structures:
- Color by Residue Position. Rainbow color scale going from blue over green to yellow and red, following the residue number.
- Color by Type. For proteins, beta sheets are blue, helices red and loops/coil gray. For nucleic acids backbone ribbons are white while the individual nucleotides are indicated in green (T/U), red (A), yellow (G), and blue (C).
- Color by Backbone Temperature. For PDB files, this is based on the b-factors for the C
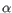 atoms (the central carbon atom in each amino acid). For structure models created with tools in the workbench, this is based on an estimate of the local model quality. The color scale goes from blue (0) over white (50) to red (100). The b-factors as well as the local model quality estimate are measures of uncertainty or disorder in the atom position; the higher the number, the higher the uncertainty.
atoms (the central carbon atom in each amino acid). For structure models created with tools in the workbench, this is based on an estimate of the local model quality. The color scale goes from blue (0) over white (50) to red (100). The b-factors as well as the local model quality estimate are measures of uncertainty or disorder in the atom position; the higher the number, the higher the uncertainty.
- Color by Entry. Each chain/molecule is assigned its own specific color.
- Custom Color. The user selects a molecule color from a palette.
