Update Sequence Attributes
Update Sequence Attributes adds or updates information in attribute columns of input sequences and outputs new sequence elements containing the modified attribute information.
Note that attributes relating to characteristics of the sequences, such as length or the start of the sequences, cannot be updated using this tool.
To launch the Update Sequence Attributes tool, go to:
Tools | Utility Tools (![]() ) | Sequences (
) | Sequences (![]() ) | Update Sequence Attributes (
) | Update Sequence Attributes (![]() )
)
The tool takes as input one sequence (![]() ) (
) (![]() ) (
) (![]() ), sequence list (
), sequence list (![]() ) (
) (![]() ), alignment (
), alignment (![]() ), or phylogenetic tree (
), or phylogenetic tree (![]() ), and is recommended when updating information for several attributes/sequences. Individual attributes can also be updated directly in the Table view of these elements.
), and is recommended when updating information for several attributes/sequences. Individual attributes can also be updated directly in the Table view of these elements.
The following options can be configured in the Options dialog (figure 27.33):
- File. Select an Excel file (
.xls/xlsx), a comma-separated text file (.csv), or a tab-separated text file (.tsv) containing attribute information. Column names are used as attribute names, so a header row is required. One column in the file must contain information that can be matched with information already present in the input sequences (see "Column to match on", below). - Column to match on. Specify the column in the attribute file to use to match each row with the relevant sequence in the input. When a value in this column matches a value in the column of the same name in the input, information from that row in the file is added to the attribute information for that sequence. Only information from specified columns will be added (see "Include columns", below.)
When matching based on sequence names, the column in the file containing the names must be called
Name. - Include columns. Select the columns in the file containing the information to be updated or added to the input as well as the column specified in the "Column to match on" field.
When the name of a column does not match an existing attribute name in the input sequences, a new attribute column will be added.
- Overwrite existing attributes. When this option is checked, existing sequence attribute values will be overwritten by values for the corresponding attributes in the attribute file. When no corresponding value is present in the attributes file, no change is made to the value in the input sequences.
When left unchecked, existing attribute values in the input sequences are not overwritten with new information from the file.
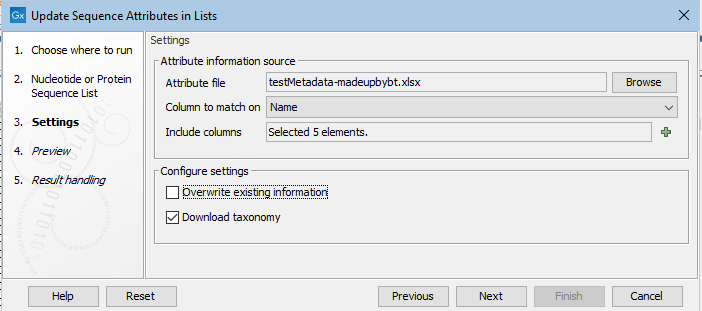
Figure 27.33: Information in the "Attributes.tsv" file will be matched with the relevant sequences based on content of the Name column in the file and in the input sequences. Six columns containing relevant attribute information have been selected.
The result of the choices made in the Options wizard step are reflected in the Preview step (figure 27.34). In the upper pane is a list of the attribute types to be updated or added, as well as the attribute to be used to match sequences with the relevant information. How particular columns will be handled is indicated in the "Content handling" column, including whether validation will be applied. The columns subject to validation checks are described later in this section.
Shown in the lower pane is a small subset of the incoming information from the attribute file, based on the choices made in the Options wizard step. Click on the "Previous" button to go back to that step if anything needs to be adjusted.
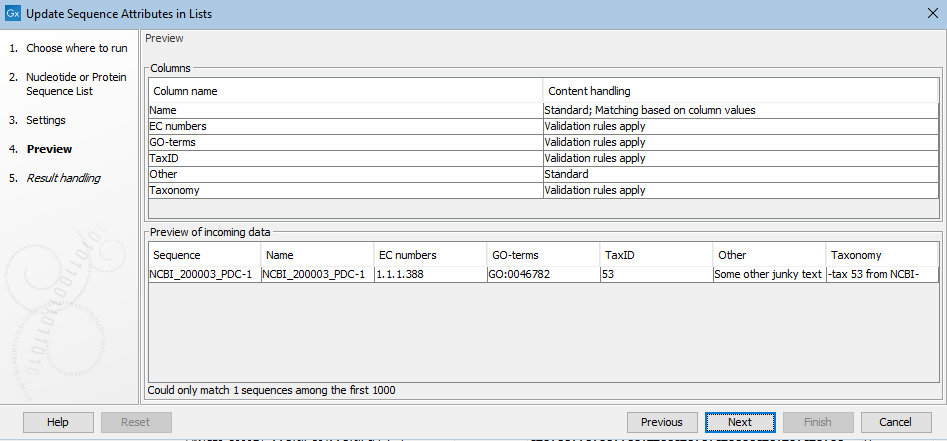
Figure 27.34: The Preview wizard steps shows information about how columns from the attribute file will be handled, and whether any problems were detected. Where validation checks are carried out, if any had failed, a yellow exclamation mark in the bottom pane would be shown for that column. Here, all entries pass. The "Other" column is not subject to validation checks. Only one sequence in the list is being updated in this example.
Column headings and value validation
Certain column names are recognized by the software and validation rules are applied to these. When the contents pass the validation checks, entries in those columns may be further processed.
In most cases, this further processing involves adding hyperlinks to online data resources. However, the contents of columns named Gene ID trigger different handling:
- The following identifiers in a Gene ID column are added as attribute values and hyperlinked to the relevant online database:
- Ensembl Gene IDs
- HUGO Gene IDs
- VFDB Gene IDs
- Any other values in a Gene ID column are added as attributes to the relevant sequences, but are not hyperlinked to an online data resource. Note that this is different to how other non-validated attribute values are handled, as described below.
- Multiple identifiers in a given cell, separated by commas, will be added as multiple Gene ID attributes for the relevant sequence. If any one of those identifiers is not recognized as one of the above types, then none will be hyperlinked.
Other columns where contents are validated are those with the headings listed below. If a value in such a column cannot be validated, it is not added nor used to update attributes.
If you wish to add information of this type but do not want this level of validation applied, use a heading other than the ones listed below.
- COG-terms. COG identifiers
- Compound AROs. Antibiotic Resistance Ontology identifiers
- Compound Class AROs. Antibiotic Resistance Ontology identifiers
- Confers-resistance-to ARO. Antibiotic Resistance Ontology identifiers
- Drug ARO. Antibiotic Resistance Ontology identifiers
- Drug Class ARO. Antibiotic Resistance Ontology identifiers
- EC numbers. EC identifiers
- GenBank accession. Genbank accession numbers
- Gene ARO. Antibiotic Resistance Ontology identifiers
- GO-terms. Gene Ontology (GO) identifiers
- KO-terms. KEGG Orthology (KO) identifiers
- Pfam domains. PFAM domain identifiers
- Phenotype ARO. Antibiotic Resistance Ontology identifiers
- PubMed IDs. Pubmed identifiers
- TIGRFAM-terms. TIGRFAM identifiers
- Virulence factor ID. Virulence Factors of Pathogenic Bacteria identifiers
