Interactive motif search
The Motifs palette (figure 18.26) in the Side Panel of sequences (![]() ) (
) (![]() ) (
) (![]() ) or sequence lists (
) or sequence lists (![]() ) (
) (![]() ) provides options for searching and managing motifs.
) provides options for searching and managing motifs.
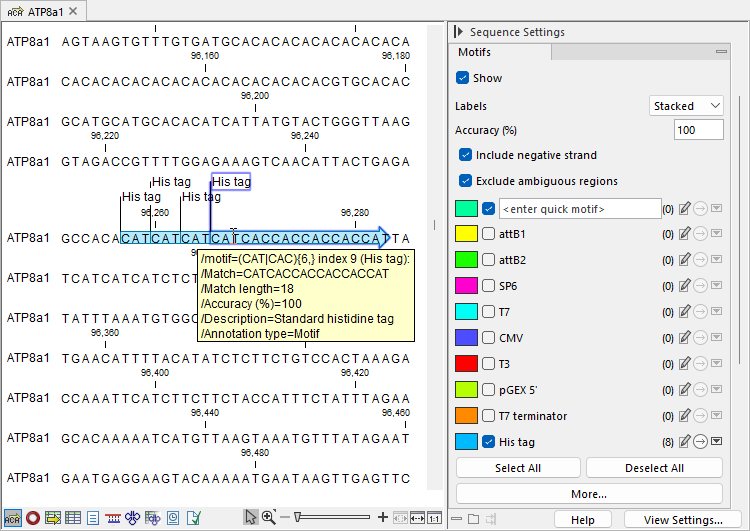
Figure 18.26: Nucleotide sequence view with the "Motifs" palette in the Side Panel expanded. Eight instances of the "His tag" motif are detected, with the checkbox in front of it checked, highlighting the matches on the sequence. Hovering the mouse over a motif region displays match information.
The following options are available:
- Show When checked, the motif options are shown in the palette, and the motifs are searched for within the sequence(s).
- Labels Control how labels for detected motifs are displayed:
- No labels
- Flag shows the motif name above the sequence
- Stacked stacks overlapping labels so that all motif names are visible.
- Accuracy (%) The percentage of residues that must match the motif. This option only impacts simple motifs. Indels are not supported.
Ambiguous characters are considered different if they do not have any residues in common. For example, N matches any nucleotide and R matches nucleotides A,G, while X matches any amino acid and Z matches amino acids E,Q.
- Include negative strand When checked, the reverse complement sequence of the motifs is also searched on the negative strand. This option is only available for nucleotide sequences.
- Exclude ambiguous region When checked, motifs are not searched for in N-regions. Additionally, when a motif matches to an N, it is treated as a mismatch.
By default, common motifs for nucleotide or protein sequences are shown. The first motif in the palette allows for a quick search by simply typing the residues to search, without the need to add a new motif. By default, this quick motif is a simple motif.
For each motif, the palette shows the motif name, or the motif itself for the quick motif, and the number of detected matches in brackets. The following actions are available for each motif:
- Check the box to highlight the detected motifs on the sequence, including their orientation. Hovering over a highlighted region shows information about the motif match (figure 18.26). For simple motifs with accuracy below 100%, matching residues are in uppercase, and non-matching residues in lowercase.
- Click the color indicator to change the highlight color for the motif.
- Click the edit icon (
 ) to open the "Edit Motif" dialog and modify the motif.
) to open the "Edit Motif" dialog and modify the motif.
- Click the right arrow icon (
 ) to navigate to the next match in the sequence. Hold Ctrl (
) to navigate to the next match in the sequence. Hold Ctrl ( on Mac) and click to navigate to the previous match.
on Mac) and click to navigate to the previous match.
- Click the down arrow icon (
 ) to display a list of all motif matches. Click a match to navigate to its location in the sequence.
) to display a list of all motif matches. Click a match to navigate to its location in the sequence.
Use Select All and Deselect All to check or uncheck all motifs in the palette.
To update the motifs shown in the palette:
- Click the More... button and select Add Motif to open the "Add Motif" dialog and add a new motif.
- Click the More... button and select Manage Motifs to open the "Manage Motifs" dialog (figure 18.27). The dialog shows the motifs currently visible in the palette in the right list, while the left list contains additional common motifs.
The following actions are available:
- Click the Browse (
 ) button to load motif lists (
) button to load motif lists ( ) into the left list.
) into the left list.
- Use the right (
 ) and left (
) and left ( ) arrows to move selected motifs between the two lists.
) arrows to move selected motifs between the two lists.
To select multiple motifs, hold Ctrl (
 on Mac) and click each desired motif. To select a range, hold Shift and click the first and last motif.
on Mac) and click each desired motif. To select a range, hold Shift and click the first and last motif.
- Use the up (
 ) and down (
) and down ( ) arrows to change the placement of the selected motif in the right list.
) arrows to change the placement of the selected motif in the right list.
- Save motifs in the right list to a new motif list by checking Save as a new motif list.
- Click the Browse (
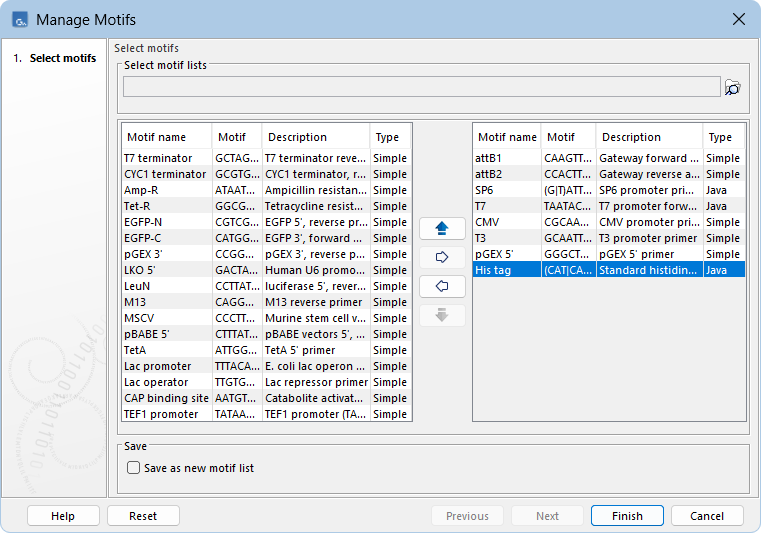
Figure 18.27: The "Manage Motifs" dialog. "His tag" is selected in the right list and can be removed from the palette by moving it to the left list using the left arrow.
