Create Motif List from Sequences
Create Motif List from Sequences takes as input sequences (![]() ) (
) (![]() ) (
) (![]() ), sequence lists (
), sequence lists (![]() ) (
) (![]() ), or alignments (
), or alignments (![]() ), and outputs a motif list (
), and outputs a motif list (![]() ) containing simple motifs created from the provided sequences or their annotations.
) containing simple motifs created from the provided sequences or their annotations.
To run the tool, go to:
Tools | General Sequence Analysis (![]() )| Motif (
)| Motif (![]() ) | Create Motif List from Sequences (
) | Create Motif List from Sequences (![]() )
)
The following options can be configured (figure 18.24):
- Motif name Defines the names of the created motifs. The following placeholders are available:
- {sequence-name} The name of the source sequence.
- {annotation-name} The name of the source annotation.
- {annotation-type} The type of the source annotation.
- {annotation-type?} The type of the source annotation if it differs from the annotation name. This is useful in combination with {annotation-name} when the name and type are identical, to avoid repetitions in the motif name.
- From sequences When checked, a motif is created for each sequence in the input, using the entire sequence as the region for the motif.
- From annotations When checked, a motif is created for each annotation in each sequence of the input, using the annotated region as the region for the motif.
- Annotation types Only annotations of the selected types are used to create motifs. If left empty, all annotation types are included.
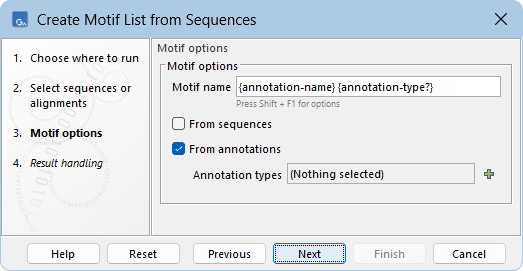
Figure 18.24: Creating a new motif list from sequences.
A simple motif is created for each region defined by the selected options, using the sequence residues within that region (figure 18.25). Each created motif contains the following information:
- The name is defined according to the Motif name option.
- The description is set from the sequence description when the motif is created from a sequence, or from the annotation description, if available, when the motif is created from an annotation.
- The "Annotation type" is set to "Motif" if the motif is created from a sequence, or to the annotation type if created from an annotation.
- The "From sequence" note indicates the source sequence, and, if applicable, the "From annotation" note indicates the source annotation used to create the motif.
- If applicable, any annotation notes present in the source annotation are included in the motif.
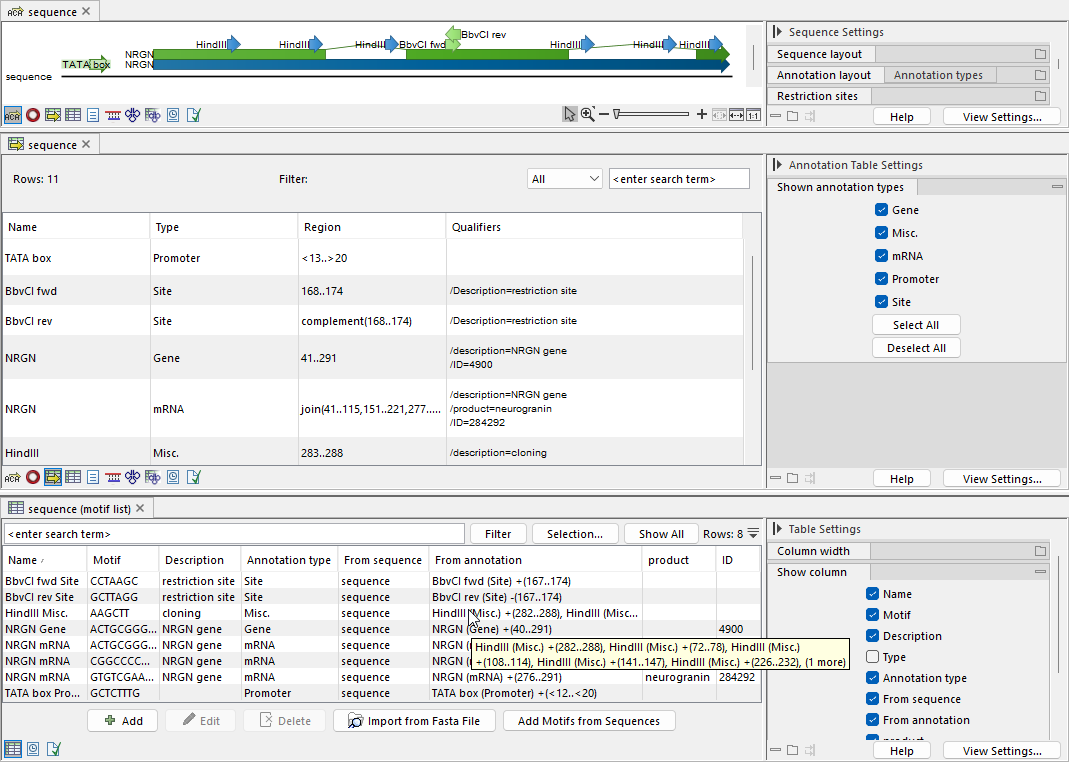
Figure 18.25: Motif list created from a sequence containing multiple types of annotations. Top: Sequence view. Middle: Sequence annotation table view. Bottom: Motif list view.
Duplicate motif collapsing
If several motifs have the same annotation type and motif sequence, they are collapsed into a single motif (figure 18.25). For the combined motif, the "Description" and each type of annotation note, including "From sequence" and "From annotation", retain up to five unique notes from the original duplicated motifs. The "Name" is set to that of the first duplicated motif.
Special handling of complex annotations
Annotations with a region that is not simple are processed as follows:
- If the annotation is spliced, meaning the region is composed of multiple joined subregions, one motif is created for each subregion. Figure 18.25 shows an example where an mRNA annotation produces several motifs.
- If the annotation has uncertain or unknown boundary points, a motif is created for the specified region. Figure 18.25 shows an example where a Promoter annotation with uncertain boundaries results in a single motif for the exact specified region.
- If the annotation is located on the minus strand, the motif is created from the residues on that strand. Figure 18.25 shows an example where two Site annotations on opposite strands produce two motifs that are reverse complements of each other.
