Create Sequence Constructs
Create Sequence Constructs builds DNA, RNA, or protein constructs by joining sequences from defined components in a specified order, generating every possible sequence combination. This can be useful for applications such as cloning, synthetic circuit design, pathway engineering, and gene-editing.
To run the tool, go to:
Tools | Cloning (![]() )| Create Sequence Constructs (
)| Create Sequence Constructs (![]() )
)
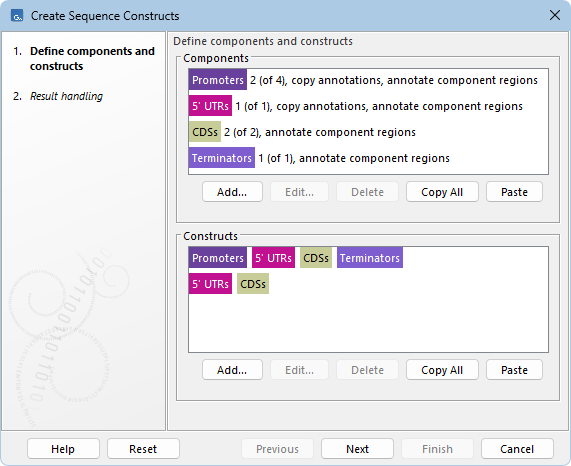
Figure 23.1: Components are defined in the top panel and constructs in the bottom panel. Here, 4 components are defined (Promoters, 5' UTRs, CDSs, Terminators) with varying numbers of included sequences. Two constructs are defined from these components. Annotations in the output indicate the component regions and, for Promoters and 5' UTRs, annotations are also copied from the input sequences.
Defining components
To add a new component, click the Add... button below the Components panel to open the "Setup component" dialog (figure 23.2) and define the component:
- Component name Used in the output.
- Sequences (Optional) One or more DNA or RNA sequences (
 ) (
) ( ) (
) ( ), or protein sequences (
), or protein sequences ( ) (
) (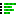 ) from the Navigation Area. Only one sequence type is allowed. DNA can be translated to protein, and protein can be reverse translated to DNA.
) from the Navigation Area. Only one sequence type is allowed. DNA can be translated to protein, and protein can be reverse translated to DNA.
The sequences are added to the table below.
- Copy annotations to constructs When checked, annotations from the selected Sequences are copied to the output.
- Annotate component regions in constructs When checked, each component region is annotated in the output using the component name as type and the sequence name as annotation name.
- Manual sequences Sequences can be manually defined in the table below.
Click the Add button to add a new row in the table and type a Name and Sequence. Manual sequences can also be deleted by selecting them and clicking the Delete button.
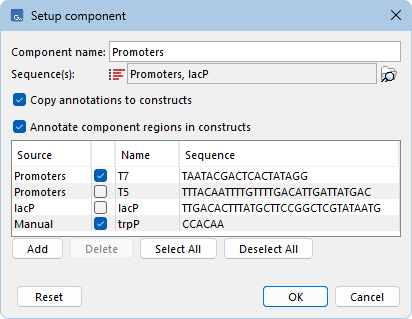
Figure 23.2: Components are set up by specifying a name and sequences.
The table in the "Setup component" dialog lists all added sequences. The Source column indicates one of the following:
- The name of the individual sequence or sequence list from which the sequence originated.
- Manual if the sequence is added manually.
Only manual sequences can be edited or deleted from the table. Sequences can be included or excluded from the component by using the checkboxes. This is useful when a sequence list contains sequences that should be excluded from the component. Select All and Deselect All can be used to check or uncheck all sequences.
To edit a component, double-click it or select it and click the Edit... button.
Selected components can be deleted by using the Delete button. To select components, hold Ctrl (![]() on Mac) and click the desired components, or hold Shift and click the first and last component in a range.
on Mac) and click the desired components, or hold Shift and click the first and last component in a range.
Each component receives a color derived from its name. This color is used both in the Constructs panel and in the annotations added to the output. Different components may share the color.
Defining constructs
To add a new construct, click the Add... button below the Constructs panel to open the "Combine components" dialog (figure 23.3). Components can be arranged in any order, reused multiple times, and not all components need to be included.
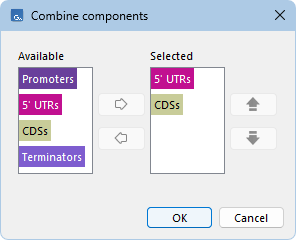
Figure 23.3: Constructs are set up by combining the defined constructs in the desired order.
To edit a construct, double-click it or select it and click the Edit... button.
Selected constructs can be deleted by using the Delete button. To select constructs, hold Ctrl (![]() on Mac) and click the desired constructs, or hold Shift and click the first and last construct in a range.
on Mac) and click the desired constructs, or hold Shift and click the first and last construct in a range.
If no constructs are defined, all components are used in their listed order to form a single construct.
Reusing components and constructs
Previously defined components and constructs can be reused in later runs.
Components and constructs can be copied in two ways:
- Use Copy all to copy the definition of either all components or all constructs defined in the tool.
- Copy both components and constructs from the History (
 ) view of a previous output (figure 23.4).
) view of a previous output (figure 23.4).
Copied definitions can be stored in a text file for later use, or pasted directly into the tool using the corresponding Paste button. Pasted items are added to those already defined.
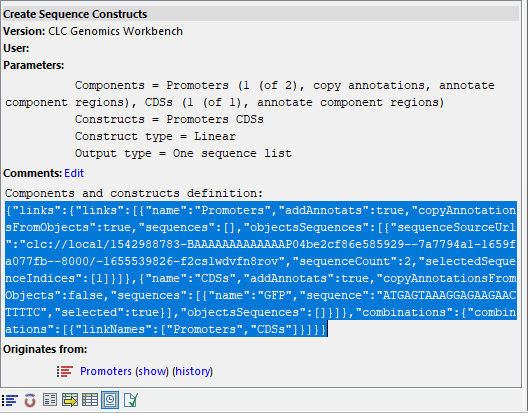
Figure 23.4: The History view of an output lists the components and constructs definitions under "Comments". These can be copied and reused in subsequent runs.
Output options
For each construct, the tool generates all possible sequences that can be formed from its components. See Create Sequence Constructs output for more details.
In the "Result handling" wizard, the following options can be configured:
- Construct type DNA and RNA constructs can be Linear or Circular. Protein constructs can only be made linear.
- Output type:
- Single sequences Each generated sequence is saved separately, with names built from the sequence names of its components.
- Multiple sequence lists All generated sequences for one construct are saved as a sequence list, named using the component names.
- One sequence list All generated sequences are saved as one sequence list.
Sequence names in lists follow the same convention as Single sequences.
Subsections
