Annotate with GFF/GTF/GVF files
Annotate with GFF/GTF/GVF files adds annotations from GFF3, GTF, or GVF files to DNA, RNA, or protein sequences. Annotations can be added to sequences stored individually (![]() ) (
) (![]() ) (
) (![]() ) or in lists (
) or in lists (![]() ) (
) (![]() ).
).
To run the tool, go to:
Tools | General Sequence Analysis (![]() )| Annotate with GFF/GTF/GVF files (
)| Annotate with GFF/GTF/GVF files (![]() )
)
The following options can be configured (figure 18.1):
- GFF/GFT/GVF files One or more GFF, GTF, or GVF files containing annotations to add to the input sequences.
Annotations from each line are added to the input sequence whose name matches the value in the first column. Rename Elements and Rename Sequences in Lists can be used to update sequence names as needed.
- Annotation type handling
- Keep types from files The annotation type is set to the type defined in the files.
- Replace with custom type The annotation type is set to the provided Custom type for all annotations.
- Annotation name handling.
- Keep names from files The annotation name is set to the value of the first available qualifier in this order: Name, Gene_name, Gene_ID, Locus_tag, ID. If none are present, the annotation type is used.
- Replace from qualifier The annotation name is set from the provided Qualifier. If the qualifier is not present, the name is as for Keep names from files.
Transcript annotations inherit the name from the gene annotation.
- Keep only one of duplicate annotations When checked, only one instance of each duplicate annotation is added to the sequences.
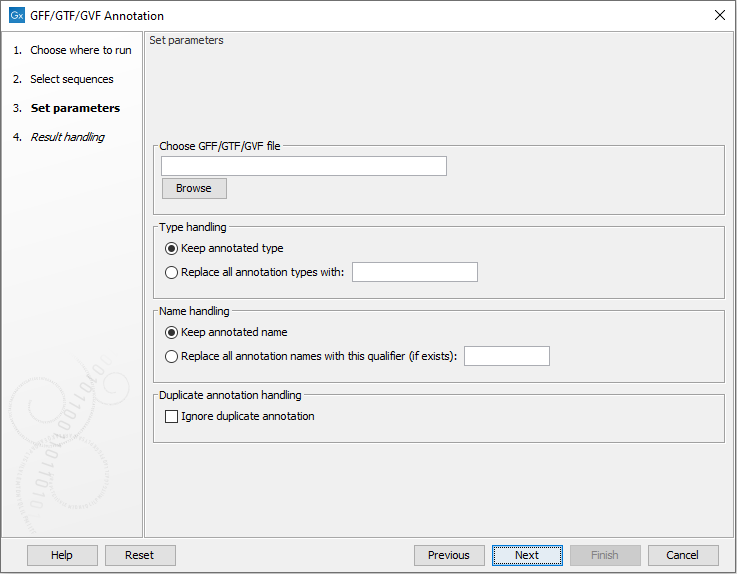
Figure 18.1: Options for annotating sequences using GFF, GTF, or GVF files.
Handling of CDS, exon, mRNA, transcript and gene annotations
Annotations of the types CDS, exon, mRNA, transcript, and gene are handled as follows:
- A gene annotation is added for each gene_id. The annotated region extends from the leftmost to the rightmost positions of all annotations with that gene_id.
- CDS annotations sharing the same transcriptID or the same parent are combined into a single CDS annotation.
- Exon annotations sharing the same mRNA or transcript parent are combined into a single mRNA or transcript annotation.
- Multiple exon annotations without a parent that share the same transcriptID are combined into a single mRNA annotation. If there is only one exon annotation for a transcriptID and there is no corresponding CDS, a transcript annotation is added instead.
Note that genes, CDS, transcripts, and mRNA annotations are linked by name only (not by position, ID, or other qualifiers).
