On-the-fly import in workflows
Most single cell data importers can be used in workflows for importing data on the fly. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Launching_workflows_individually_in_batches.html for general information about on-the-fly import. This section provides information specific to the single cell data importers.
Note that when importing data on the fly, the Preview cells section, see Cell format in importers, is not available.
Importing multiple datasets
When importing multiple datasets, the same import options are used for all datasets. For example, if the files to be imported do not share the same Cell format and/or Sample, they need to be imported separately before running the workflow.
Most importers use a single file for importing data. To import multiple datasets, select all the files to be imported.
Some importers allow optional additional files, and others require several files. For example, Import Peak Count Matrix in Cell Ranger HDF5 format imports the data from an HDF5 file, and can optionally import nearby genes and/or transcription factors from additional files, while Import Expression Matrix in MEX format requires at least three files: barcodes, features and matrix files.
Importers that accept more than one file have the following additional options when used in a workflow:
- Use archive file. Enables import of an archive file instead of individual files.
- Archive file. The archive file to be imported. Hover the mouse cursor over the option to see a tooltip with a description of the files the importer will use from the archive (figure 4.15).
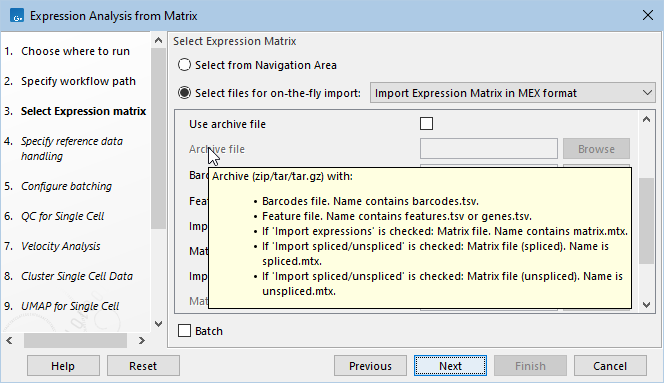
Figure 4.15: Hovering the mouse cursor over the 'Archive file' option reveals a tooltip with a description of the files the importer will use from the archive.
To import multiple datasets using such importers, check the Use archive file option and select all the archive files to be imported.
Batching
When importing multiple datasets, batching can be used to analyze the datasets separately.
Note that most template workflows provided by the CLC Single Cell Analysis Module include an Iterate control flow element, which automatically handles batching when multiple files are imported.
Batching can sometimes be disallowed (figure 4.16).
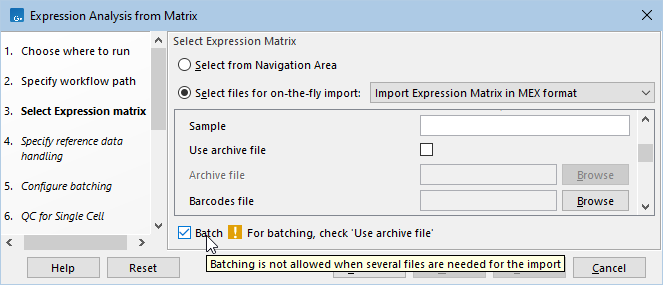
Figure 4.16: Hovering the mouse cursor over the 'Batch' option reveals a tooltip. When batching is disallowed, the tooltip and an info message offer a description of why batching is disallowed and what is required for batching.
For more information on running workflows in batch mode, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Running_workflows_in_batch_mode.html.
