Barcodes from names
In some protocols, the cell barcode is not included in the read. For instance, the Illumina sample index can be used as a barcode:
- For Smart-seq2, the cell barcode is the sample index.
- For Parse Biosciences, the sample index can optionally be used as the fourth barcode, see Parse Biosciences.
The cell barcode can be extracted from the name of the reads and/or input sequence list by adjusting the options in the Barcodes from names dialog (figure 6.3):
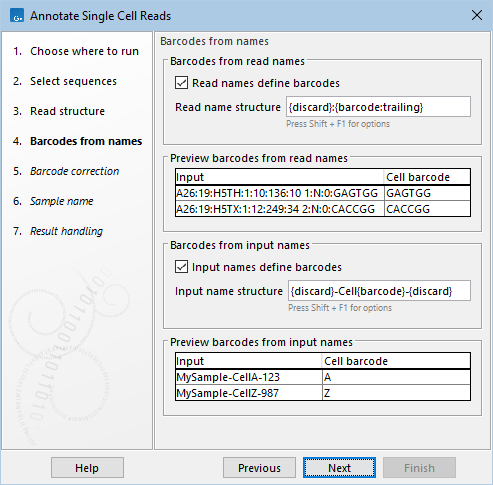
Figure 6.3: Extracting cell barcodes from the read and input names. The two previews show how the names are parsed into cell barcodes.
- Read/Input names define barcodes. When checked, cell barcodes are extracted from the read/input names, according to the structure configured in Read/Input name structure.
- Read/Input name structure. Specify how to extract cell barcodes from the read/input names. The cell barcode does not have to consist of nucleotides. A combination of placeholders and text can be used (figure 6.3) to extract the part(s) of the name containing the cell barcode. Hover the mouse cursor over the field to see a tooltip with several examples. Simultaneously pressing Shift and F1 displays all available placeholders.
The Barcodes from names dialog has two previews, displaying how the read/input names are parsed into the cell barcode (figure 6.3). These previews help ensure that the name structure matches the input. If the configured structure is invalid, the preview may fail to determine the cell barcode.
