Compare Cell Clonotypes
Compare Cell Clonotypes contrasts properties, such as diversity and similarity, of the immune repertoires identified for groups of cells, as determined by the sample or, when available, through Cell Clusters (Compare Cell Clonotypes is available from:
Tools | Single Cell Analysis (![]() ) | Immune Repertoire (
) | Immune Repertoire (![]() ) | Compare Cell Clonotypes (
) | Compare Cell Clonotypes (![]() )
)
The tool takes a TCR Cell Clonotypes (![]() ) or BCR Cell Clonotypes (
) or BCR Cell Clonotypes (![]() ) element as input. If the clonotypes to be compared are found in different elements, these can be combined using Combine Cell Clonotypes, see Combine Cell Clonotypes for details.
) element as input. If the clonotypes to be compared are found in different elements, these can be combined using Combine Cell Clonotypes, see Combine Cell Clonotypes for details.
The following options can be adjusted (figure 13.5):
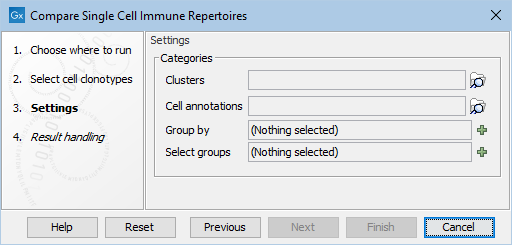
Figure 13.5: The default options in the dialog of Compare Cell Clonotypes. Note that "Group by" needs to contain at least one value before proceeding. This can always be set to "Sample".
- Clusters and Cell annotations (Optional). Clusters accepts Cell Clusters (
 ) and Cell annotations accepts Cell Annotations (
) and Cell annotations accepts Cell Annotations ( ). These can be created within the CLC Single Cell Analysis Module for scRNA-Seq data, but they can also be imported, see Import Cell Annotations and Import Cell Clusters for details.
). These can be created within the CLC Single Cell Analysis Module for scRNA-Seq data, but they can also be imported, see Import Cell Annotations and Import Cell Clusters for details.
- Group by. Any categories from the supplied Cell Clusters or Cell Annotations. "Sample" can be additionally chosen, even if no Cell Clusters or Cell Annotations are provided.
If Cell Annotations contained a category `Infection' with values `Pre' and `Post', and another category `Individual' with values `Ind1' and `Ind2', then selecting Group by = Infection, Individual would give groups `Pre - Ind1', `Pre - Ind2', `Post - Ind1' and `Post - Ind2'.
The chosen options need to induce at least two groups of cells. Otherwise, the tool will fail with a relevant message.
Note that category from Cell Annotations that only contain non-integer numerical data are not supported.
- Select groups (Optional). This can be supplied to reduce the number of groups of cells in the outputs to only those of interest, or to control the order in which the groups are shown. For example, if the aim is to investigate the infection effect in individual 1, the `Pre - Ind1' and `Post - Ind1' groups can be selected. If left empty, all groups will be used.
Subsections
