Normalize Single Cell Data
Normalize Single Cell Data transforms expression data to remove the effect of sequencing depth and, optionally, the effect of batch factors. Data normalization is essential prior to downstream analysis. Batch correction is performed by choosing one batch, corresponding to one factor, as the 'baseline', and applying transformations to the remaining batches such that they resemble the baseline. Batch correction is appropriate only if a batch effect is evident in a Dimensionality Reduction Plot.
The tool is available from:
Tools | Single Cell Analysis (![]() ) | Gene Expression (
) | Gene Expression (![]() ) | Cell Preparation (
) | Cell Preparation (![]() ) | Normalize Single Cell Data (
) | Normalize Single Cell Data (![]() )
)
Normalize Single Cell Data takes at least one Expression Matrix (![]() ) / (
) / (![]() ) as input, and produces a single Expression Matrix (
) as input, and produces a single Expression Matrix (![]() ) / (
) / (![]() ) as output, as well as an optional report. If multiple inputs are provided, the output will only contain the genes that are present in all inputs. The tool disregards any normalization already stored on its inputs.
) as output, as well as an optional report. If multiple inputs are provided, the output will only contain the genes that are present in all inputs. The tool disregards any normalization already stored on its inputs.
The following options can be configured in the Batch correction dialog (figure 7.23):
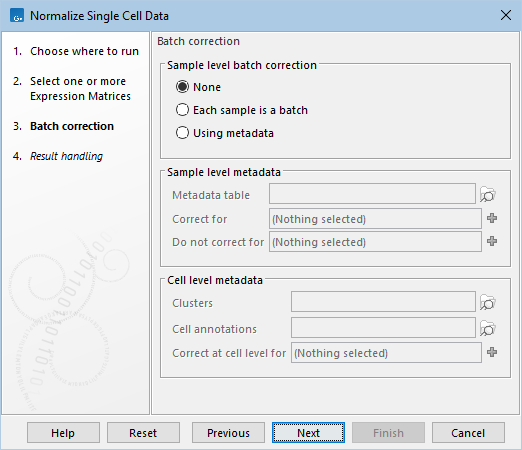
Figure 7.23: The default settings in the Batch correction dialog.
- None. Batch correction is not applied, but expression data is transformed to remove the effect of sequencing depth.
- Each sample is a batch. Each batch consists of one sample. This is suitable when systematic changes in expression are expected across samples and are irrelevant to downstream analysis, such as when combining samples of the same tissue created by different investigators.
- Using metadata.
- Sample level metadata. Batches can be specified at the level of inputs. This requires that multiple inputs are provided.
- Metadata table. A CLC Metadata Table defining how inputs are grouped into batches.
- Correct for. Columns of the metadata table corresponding to batch factors, see The output of Normalize Single Cell Data.
- Do not correct for. Columns of the metadata table. When used, the tool explicitly models the effect of these factors on expression, helping to prevent batch correction from inadvertently removing variation attributable to these factors. The tool will warn if all variation due to the Do not correct for factors is removed in at least one sample, meaning that the Correct for and Do not correct for factors are confounded.
A typical use case for sample level metadata arises when combining samples of the same tissue prepared by different investigators, each handling multiple samples. Here one would set 'Correct for = investigator'. If each investigator prepared a mix of treated and control samples, then one would set 'Correct for = investigator' and 'Do not correct for = Treatment/Control'.
- Cell level metadata. Batches can be specified at the level of individual cells.
- Clusters and Cell annotations. Clusters accepts Cell Clusters (
 ) and Cell annotations accepts Cell Annotations (
) and Cell annotations accepts Cell Annotations ( ). At least one element must be supplied.
). At least one element must be supplied.
- Correct at cell level for. Categories in the clusters and/or annotations corresponding to batch factors. Cells assigned to a cluster/annotation with fewer than 20 cells, or lacking a cluster/annotation, are categorized under an "Unknown" factor. Numerical categories are not supported, so metrics like 'Mitochondrial counts (%)' cannot be regressed out - a practice that is generally not recommended [Germain et al., 2020].
A typical use case for cell level metadata arises when correcting for cell cycle effects.
- Clusters and Cell annotations. Clusters accepts Cell Clusters (
Care must be taken when defining the batch factors, as it is easy to over-parameterize the model, see The output of Normalize Single Cell Data.
- Sample level metadata. Batches can be specified at the level of inputs. This requires that multiple inputs are provided.
Subsections
- When is batch correction appropriate?
- The output of Normalize Single Cell Data
- The Normalize Single Cell Data algorithm
