Importing reads
The ATAC or GEX reads or both can be imported with on-the-fly imports when running the workflow. This section will describe an example for the reads shown in figure 19.14.
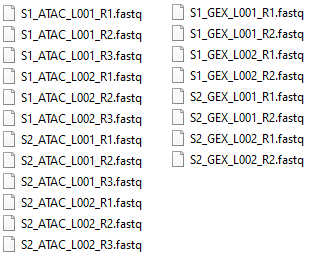
Figure 19.14: FASTQ files for two samples with two lanes for both ATAC and GEX reads
The two sets of reads must be matched with metadata as shown in figure 19.15 and 19.16. Both tables have a common column "Sample" which will be used for matching. Note that the import of 10x ATAC reads must be configured with "Custom reads options" set to "R1,R2 R3" as shown in figure 19.17.
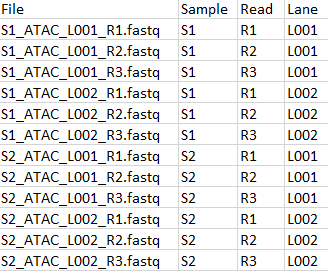
Figure 19.15: Metadata for ATAC FASTQ files
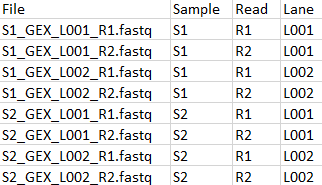
Figure 19.16: Metadata for GEX FASTQ files
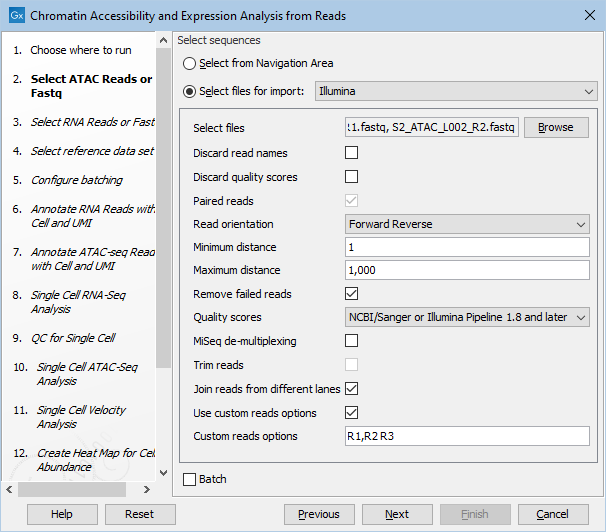
Figure 19.17: Import settings for 10x ATAC FASTQ files
The two sets of reads will be grouped in two batches as shown in figure 19.18.
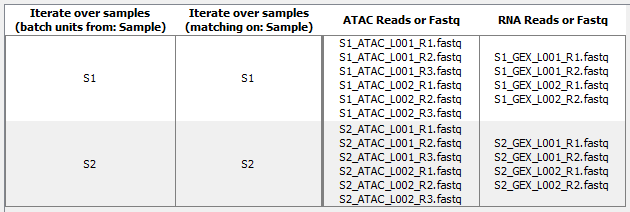
Figure 19.18: Batch overview for ATAC and GEX FASTQ files
