Reference data management
Template workflows delivered by the CLC Single Cell Analysis Module are configured to use QIAGEN Reference Sets, making them simple to launch while helping ensure that the same reference data is used consistently. This reference data can be easily obtained using the Reference Data Manager in the CLC Genomics Workbench. Reference data for a specific workflow can also be downloaded via workflow launch wizards. These features are described in detail in the section about QIAGEN Sets.
QIAGEN Sets for single cell analyses
Single cell data sets for Human and Mouse are called Single Cell hg38 (Ensembl) and Single Cell Mouse (Ensembl), respectively, and are available under the QIAGEN Sets tab of the Reference Data Manager (figure 2.1).
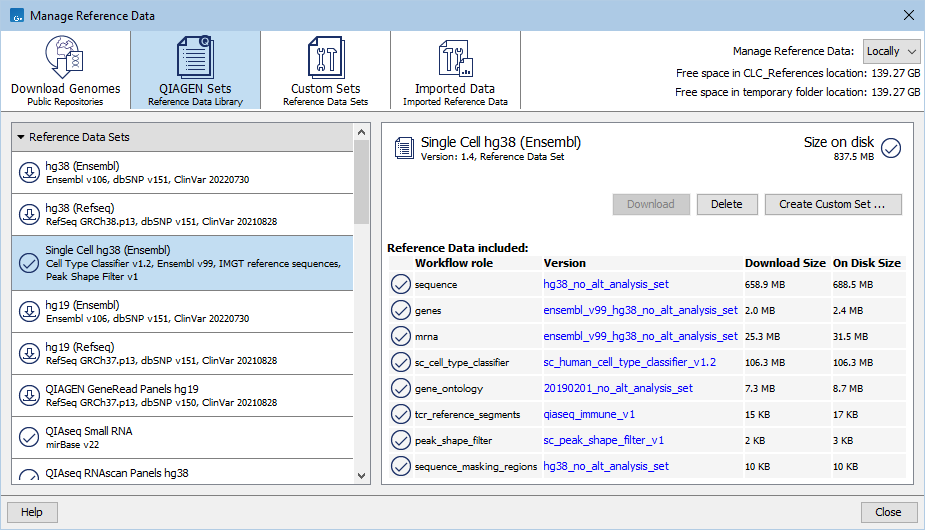
Figure 2.1: A Single Cell reference data set viewed via the QIAGEN Sets tab of the Reference Data Manager.
The reference data sets contain:
- The reference sequence, gene track, and mRNA track, used for mapping scRNA-Seq data.
- A pre-trained classifier with cell types from QIAGEN Cell Ontology to use when predicting cell types or training with more cell types.
- A gene ontology that can be used together with differential expression data to analyze GO terms.
- Reference V (variable), D (diversity), J (joining) and C (constant) gene segments, used for mapping scTCR-Seq data.
- A peak shape filter used for calling scATAC-Seq peaks.
Further detail about working QIAGEN reference data is provided in QIAGEN Sets.
Subsections
