Choosing barcodes to retain
In the dialog for Count-based filters, a list of barcodes to be retained as cells can be specified. The desired barcodes can be obtained from the plots in the report:
- Double-click on the desired plot in the report to open it.
- Change to the plot table view (
 ).
).
- Use standard table filtering tools to list the barcodes of interest (figure 7.20). See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Filtering_tables.html for details.
- Copy the barcodes and paste them in the Barcodes to retain field.
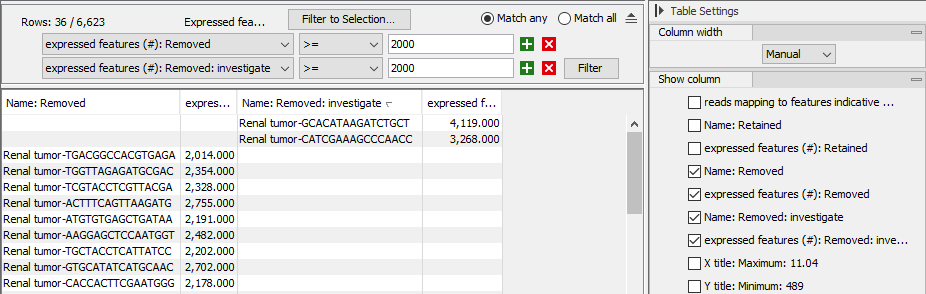
Figure 7.20: Table view of the "Expressed features vs features indicative of low quality" plot from figure 7.16. The table is filtered to only show barcodes that have been removed and have at least 2,000 expressed features.
When QC for Single Cell is run with retaining these barcodes, it will produce a report that highlights the retained barcodes as known cells, as shown in figure 7.21.
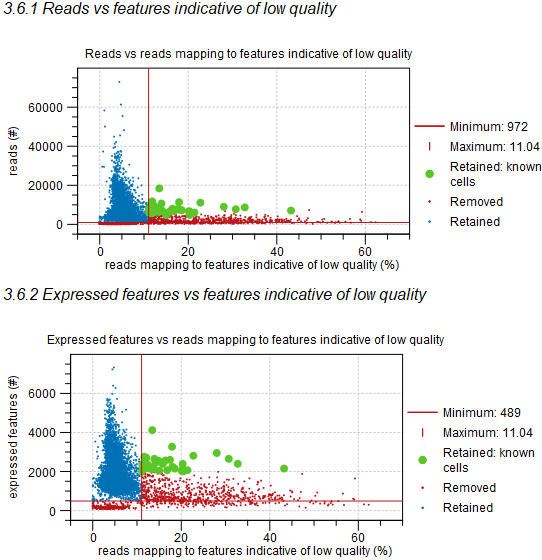
Figure 7.21: The percentage of reads mapped to features indicative of low quality vs the total number of reads (top) and expressed features (bottom). Barcodes in red have been removed and those in blue have been retained. The thresholds for removing barcodes are shown as horizontal and vertical red lines. Barcodes highlighted in green have been retained as specified known cells.
Note that this option also affects the Doublets filter. All barcodes to be retained as cells will be retained regardless of their doublet score. Such barcodes are highlighted in the doublet score relations plots.
