Configuring the batch units for Chromatin Accessibility Analysis from Reads
Consider the FASTQ files shown in figure 19.6.
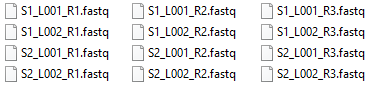
Figure 19.6: Example of twelve FASTQ files for paired reads split in three files each. They originate from two libraries with two lanes each.
The files can be automatically imported during workflow execution by choosing "Select files for on-the-fly import", selecting the Illumina importer and enabling "Paired reads" and "Join reads from different lanes". Note that for 10x ATAC reads "Use custom reads options" must be checked and "Custom reads options" must be set to "R1,R2 R3", as shown in figure 19.7. Selecting "Use organization of input data" when defining the batch units will lead to the input files being grouped in two libraries, as shown in figure 19.8.
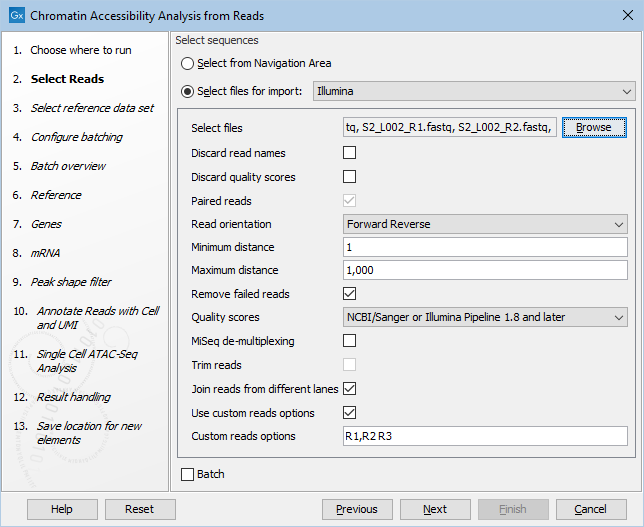
Figure 19.7: Example of importing on-the-fly twelve FASTQ files for paired reads, originating from multiple lanes and two libraries.
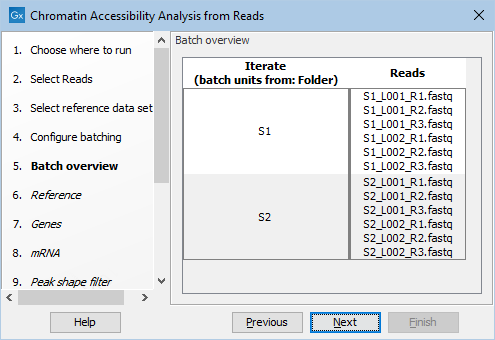
Figure 19.8: Batch overview when importing the FASTQ files and choosing "Use organization of input data".
The FASTQ files can also be imported manually and used for the workflow execution.
