Empty droplets filter
In droplet-based data, barcodes can correspond to droplets containing one cell, more cells or no cells at all. In the first dialog of QC for Single Cell, the Empty droplets filter can be enabled and customized to remove the droplets that are detected to not contain any cells. This filter should be skipped for single-cell protocols that are not droplet-based.
Note that each droplet is assigned one barcode and these terms can be used interchangeably for droplet-based protocols.
Non-zero counts in empty droplets are obtained from ambient (i.e., extracellular) RNA, that can be captured and sequenced during the protocol. Sequenced empty droplets contain significantly fewer reads, and this can be seen as a sharp transition in the rank plot, shown in figure 7.9.
Droplets can be classified in three categories (see figure 7.9):
- Ambient: removed droplets that have a low number of reads which are assumed to only contain ambient RNA.
- Cells: retained droplets that have a high number of reads.
- The remaining droplets with an intermediate number of reads can either be cells with low RNA content or empty droplets, and this cannot be determined purely based on the number of reads.
Droplets with a low number of reads are removed as ambient droplets. The threshold for this is usually obtained automatically from the histogram of number of reads, see Cell calling for details.
Droplets with a high number of reads are automatically retained as cell-containing droplets. The threshold for this is usually obtained from the automatically inferred knee from the rank plot, see figure 7.9 and Cell calling for details.
To detect cells with low RNA content, first an ambient RNA profile is estimated from the ambient droplets. The remaining droplets with an intermediate number of reads can be tested against this profile and are assigned simulation-based FDR-corrected p-values, from which non-empty droplets are identified.
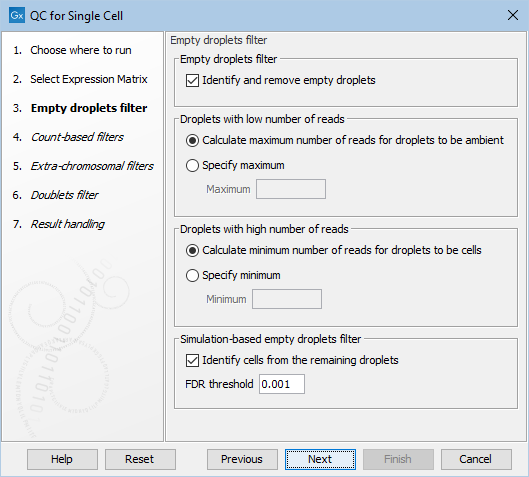
Figure 7.5: The default settings in the Empty droplets filter dialog.
The following options can be adjusted in the Empty droplets filter dialog (figure 7.5):
- Identify and remove empty droplets. Enables filtering of the empty droplets. This should be unchecked for single-cell protocols that are not droplet-based.
- Droplets with low number of reads. Droplets with a total number of reads below a threshold are removed as ambient droplets. The threshold can be calculated automatically (see Cell calling for details) by choosing Calculate maximum number of reads for droplets to be ambient, or can be specified manually in the Maximum parameter by choosing Specify maximum.
- Droplets with high number of reads. Droplets with a total number of reads above a threshold are retained as cell-containing droplets. The threshold can be calculated automatically (see Cell calling for details) by choosing Calculate minimum number of reads for droplets to be cells, or can be specified manually in the Minimum parameter by choosing Specify minimum.
- Identify cells from the remaining droplets. Enables the simulation-based detection of cells with low RNA content.
- FDR threshold. Droplets with FDR-corrected p-values larger than this are removed as empty droplets.
The generated rank plot and summary (see figures 7.9 and 7.10) can be used to identify when the automatic thresholds are not suitable and manual thresholds are required.
After applying the Empty droplets filter, only droplets that are identified as non-empty are retained for the remaining filters (see Count-based and extra-chromosomal filters). Note that this filter does not concern the quality of the retained cells. The Empty droplets filter already removes cells with low number of reads, or, by association, low number of expressed features, and enabling the Count-based filters is not strictly necessary. The Extra-chromosomal filters provide the most additional benefit in this situation.
For removing droplets containing more than one cell, the Doublets filter can be used, see Doublets filter.
