Convert Clonotypes to Cell Annotations
The Convert Clonotypes to Cell Annotations tool is available from:
Tools | Single Cell Analysis (![]() ) | Immune Repertoire (
) | Immune Repertoire (![]() ) | Convert Clonotypes to Cell Annotations (
) | Convert Clonotypes to Cell Annotations (![]() )
)
The tool takes a TCR Cell Clonotypes (![]() ) or BCR Cell Clonotypes (
) or BCR Cell Clonotypes (![]() ) element as input and produces a single Cell Annotations (
) element as input and produces a single Cell Annotations (![]() ) element summarizing the clonotypes.
) element summarizing the clonotypes.
The output contains multiple categories that summarize the primary clonotypes for each barcode (see Primary and secondary clonotypes). For converting secondary clonotypes, run first Filter Cell Clonotypes with `Multiple clonotypes' set to `Retain secondary', see Filter Cell Clonotypes for details.
The cells can be colored by any of the available categories in a Dimensionality Reduction Plot (see Dimensionality reduction) obtained from matched scRNA-Seq data for the same cells (see figure 13.7).
|
The cells in the Dimensionality Reduction Plot and those in the Cell Annotations need to have the same sample name. Ideally, it should be ensured that these share the sample name as a first step in the analysis pipeline, when running the Annotate Single Cell Reads tool (see Annotate Single Cell Reads), or when importing the Cell Clonotypes element (see Import Cell Clonotypes). If this has not been done, the sample name can be updated using the Update Single Cell Sample Name tool (see Update Single Cell Sample Name). |
The Cell Annotations (![]() ) element contains the Clone size for each barcode: the number of barcodes it shares its primary clonotype with. Let us consider the following example with TCR clonotypes, where each column represents one barcode, and the rows identify the TRA and TRB clonotypes by name:
) element contains the Clone size for each barcode: the number of barcodes it shares its primary clonotype with. Let us consider the following example with TCR clonotypes, where each column represents one barcode, and the rows identify the TRA and TRB clonotypes by name:
| B1 | B2 | B3 | |
|---|---|---|---|
| Primary TRA clonotype | TRA-1 | TRA-2 | TRA-1 |
| Primary TRB clonotype | TRB-1 | TRB-2 | None |
| Secondary TRA clonotype | TRA-2 | None | None |
| Secondary TRB clonotype | TRB-2 | None | None |
For each identified chain, the Cell Annotations (![]() ) element additionally contains the following categories with information about the primary clonotype (see figure 13.6):
) element additionally contains the following categories with information about the primary clonotype (see figure 13.6):
- productive status;
- V, D, J and C segments;
- CDR3 length;
- the CDR3 amino acid sequence;
- the number of UMIs and reads supporting the clonotype (only for single chains).
The possible identified chains are:
- chain combinations TRA + TRB and TRG + TRD for T cells;
- individual chains TRA, TRB, TRG and TRD for T cells;
- chain combinations IGH + IGK and IGH + IGL for B cells;
- individual chains IGH, IGK and IGL for B cells.
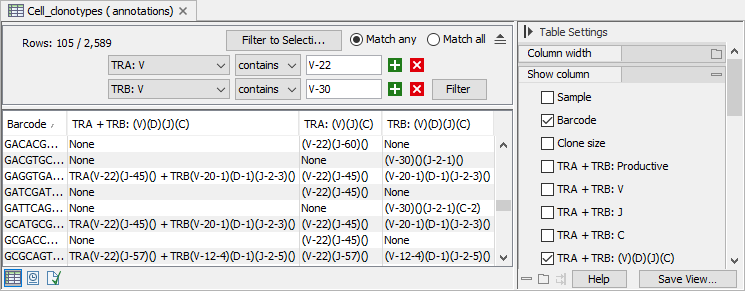
Figure 13.6: View of Convert Clonotypes to Cell Annotations output from a TCR Cell Clonotypes, filtered to specific V segments and where the segments are shown for the primary clonotypes and the TRA + TRB, TRA and TRB chains. Not all barcodes have identified clonotypes for both TRA and TRB chains.
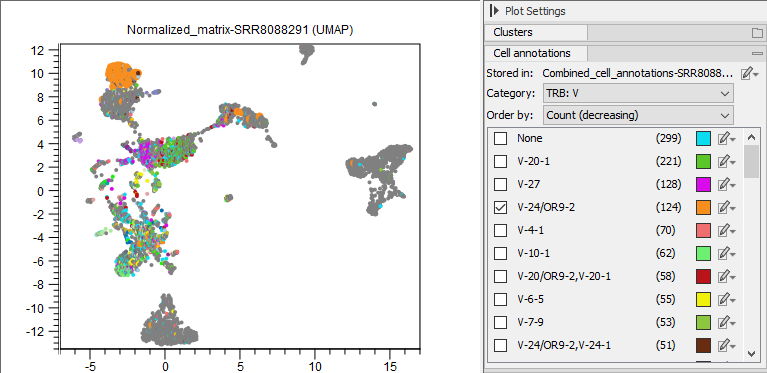
Figure 13.7: UMAP view of scRNA-Seq data, where cells are colored by the V segment from the TRB chain.
