Restriction Site Analysis
Besides the dynamic restriction sites, you can do a more elaborate
restriction map analysis with more output format using the Toolbox:
Toolbox | Molecular Biology Tools (![]() ) | Cloning and Restriction Sites (
) | Cloning and Restriction Sites (![]() )| Restriction Site Analysis (
)| Restriction Site Analysis (![]() )
)
You first specify which sequence should be used for the analysis. Then define which enzymes to use as basis for finding restriction sites on the sequence (see Manage enzymes).
In the next dialog, you can use the checkboxes so that the output of the restriction map analysis only include restriction enzymes which cut the sequence a specific number of times (figure 18.10).
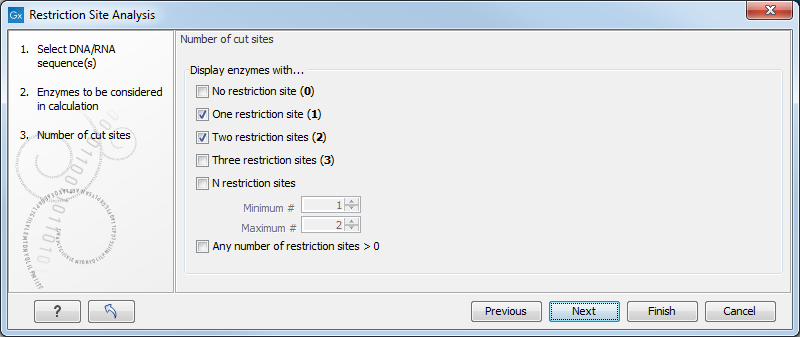
Figure 18.10: Selecting number of cut sites.
The default setting is to include the enzymes which cut the sequence one or two times, but you can use the checkboxes to perform very specific searches for restriction sites, for example to find enzymes which do not cut the sequence, or enzymes cutting exactly twice.
The Result handling dialog (figure 18.11) lets you specify how the result of the restriction map analysis should be presented.
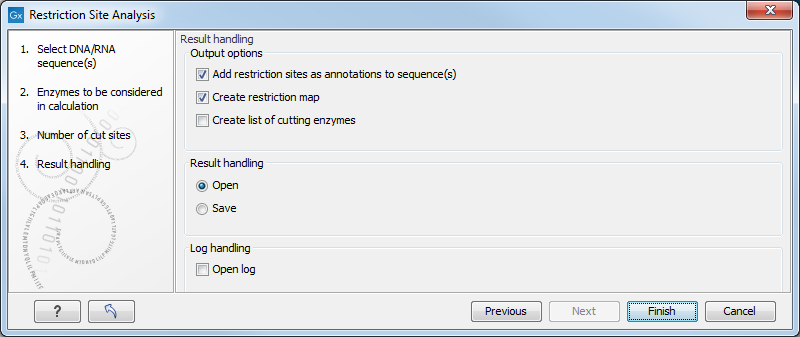
Figure 18.11: Choosing to add restriction sites as annotations or creating a restriction map.
Add restriction sites as annotations to sequence(s)
. This option makes it possible to see the restriction sites on the sequence (see figure 18.12) and save the annotations for later use.
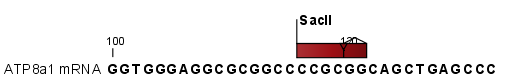
Figure 18.12: The result of the restriction analysis shown as annotations.
Create restriction map
. When a restriction map is created, it can be shown in three different ways:- As a table of restriction sites as shown in figure 18.13. If more than one sequence
were selected, the table will include the restriction sites of all the sequences. This makes it easy to compare the result of
the restriction map analysis for two sequences.
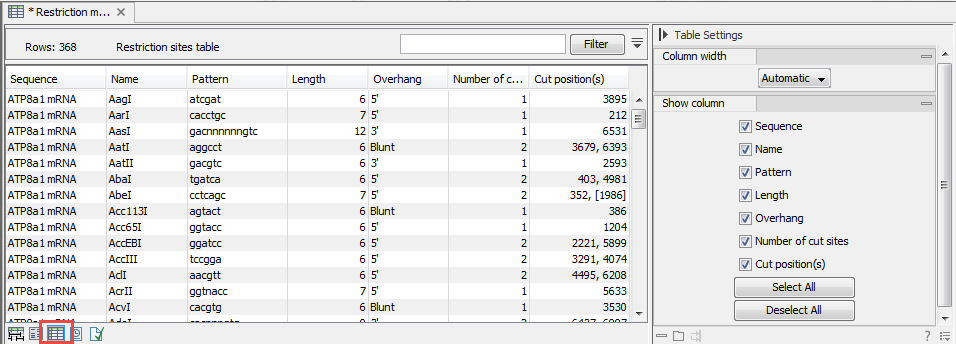
Figure 18.13: The result of the restriction analysis shown as a table of restriction sites.Each row in the table represents a restriction enzyme. The following information is available for each enzyme:
- Sequence. The name of the sequence which is relevant if you have performed restriction map analysis on more than one sequence.
- Name. The name of the enzyme.
- Pattern. The recognition sequence of the enzyme.
- Length. the restriction site length.
- Overhang. The overhang produced by cutting with the enzyme (3', 5' or Blunt).
- Number of cut sites.
- Cut position(s). The position of each cut.
- [] If the enzyme's recognition sequence is on the negative strand, the cut position is put in brackets.
- () Some enzymes cut the sequence twice for each recognition site, and in this case the two cut positions are surrounded by parentheses.
- As a table of fragments which shows the sequence fragments that would be the result of cutting the sequence with the selected enzymes (see figure18.14). Click the Fragments button
(
 ) at the bottom of the view.
) at the bottom of the view.
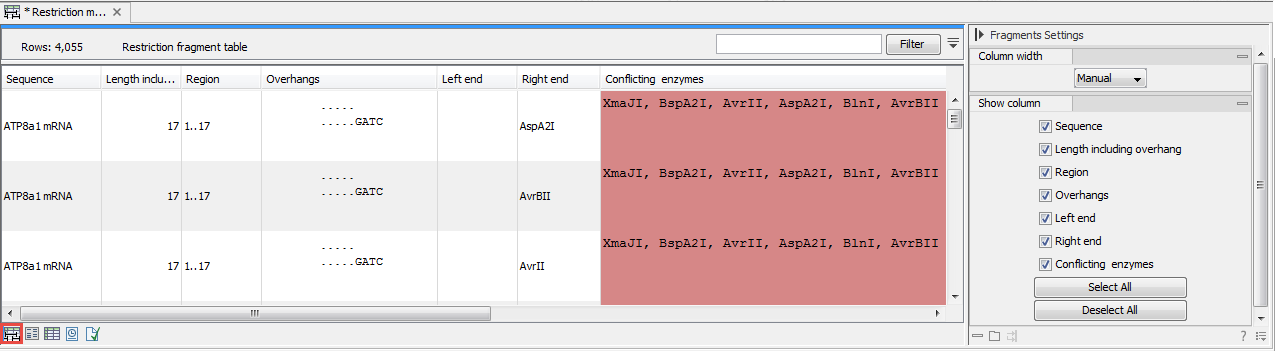
Figure 18.14: The result of the restriction analysis shown as table of fragments.Each row in the table represents a fragment. If more than one enzyme cuts in the same region, or if an enzyme's recognition site is cut by another enzyme, there will be a fragment for each of the possible cut combinations. Furthermore, if this is the case, you will see the names of the other enzymes in the Conflicting Enzymes column.
The following information is available for each fragment.
- Sequence. The name of the sequence which is relevant if you have performed restriction map analysis on more than one sequence.
- Length including overhang. The length of the fragment. If there are overhangs of the fragment, these are included in the length (both 3' and 5' overhangs).
- Region. The fragment's region on the original sequence.
- Overhangs. If there is an overhang, this is displayed with an abbreviated version of the fragment and its overhangs. The two rows of dots (.) represent the two strands of the fragment and the overhang is visualized on each side of the dots with the residue(s) that make up the overhang. If there are only the two rows of dots, it means that there is no overhang.
- Left end. The enzyme that cuts the fragment to the left (5' end).
- Right end. The enzyme that cuts the fragment to the right (3' end).
- Conflicting enzymes. If more than one enzyme cuts at the same position, or if an enzyme's recognition site is cut by another enzyme, a fragment is displayed for each possible combination of cuts. At the same time, this column will display the enzymes that are in conflict. If there are conflicting enzymes, they will be colored red to alert the user. If the same experiment were performed in the lab, conflicting enzymes could lead to wrong results. For this reason, this functionality is useful to simulate digestions with complex combinations of restriction enzymes.
If views of both the fragment table and the sequence are open, clicking in the fragment table will select the corresponding region on the sequence.
- As a virtual gel simulation which shows
the fragments as bands on a gel (see figure 18.40). For more information about gel electrophoresis, see Gel
electrophoresis.
