Expression value
Please note that reads that map outside genes are counted as intergenic hits only and thus do not contribute to the expression values. If a read maps equally well to a gene and to an inter-genic region, the read will be placed in the gene.
The expression values are created on two levels as two separate result files: one for genes and one for transcripts (if the "Genome annotated with genes and transcripts" is selected in figure 28.4). The content of the result files is described in RNA-Seq results.
The Expression value parameter describes how expression per gene or transcript can be defined in different ways on both levels:
- Total counts. When the reference is annotated with genes only, this value is the total number of reads mapped to the gene. For un-annotated references, this value is the total number of reads mapped to the reference sequence. For references annotated with transcripts and genes, the value reported for each gene is the number of reads that map to the exons of that gene. The value reported per transcript is the total number of reads mapped to the transcript.
- Unique counts. This is similar to the above, except only reads that are uniquely mapped are counted (read more about the distribution of non-specific matches in Defining mapping options for RNA-Seq).
- TPM. (Transcripts per million). This is computed as
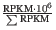 , where the sum is over the RPKM values of all genes/transcripts.
, where the sum is over the RPKM values of all genes/transcripts.
- RPKM. This is a normalized form of the "Total counts" option (see more in Definition of RPKM).
Please note that all values are present in the output. The choice of expression value only affects how Expression Tracks are visualized in the track view but the results will not be affected by this choice as the most appropriate expression value is automatically selected for the analysis being performed: for detection of differential expression this is the "Total counts" value, and for the other tools this is a normalized and transformed version of the "Total counts" as described below.
