Join alignments
CLC Genomics Workbench can join several alignments into one. This feature can for example be used to construct "supergenes" for phylogenetic inference by joining alignments of several disjoint genes into one spliced alignment. Note, that when alignments are joined, all their annotations are carried over to the new spliced alignment.
Alignments can be joined by:
Toolbox | Classical Sequence Analysis (![]() ) | Alignments and Trees (
) | Alignments and Trees (![]() )|
Join Alignments (
)|
Join Alignments (![]() )
)
This opens the dialog shown in figure 20.13.
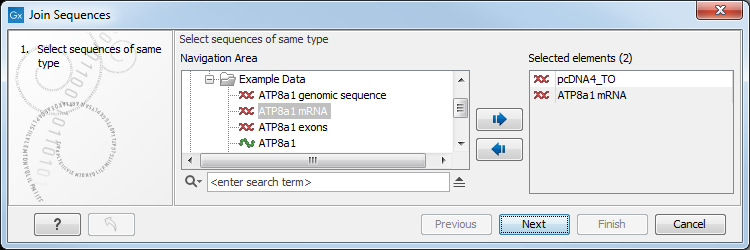
Figure 20.13: Selecting two alignments to be joined.
If you have selected some alignments before choosing the Toolbox action, they are now listed in the Selected Elements window of the dialog. Use the arrows to add or remove alignments from the selected elements. In this example seven alignments are selected. Each alignment represents one gene that have been sequenced from five different bacterial isolates from the genus Nisseria. Clicking Next opens the dialog shown in figure 20.14.
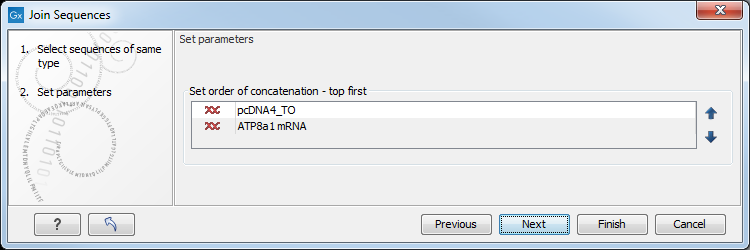
Figure 20.14: Selecting order of concatenation.
To adjust the order of concatenation, click the name of one of the alignments, and move it up or down using the arrow buttons.
The result is seen in the lower part of figure 20.15.
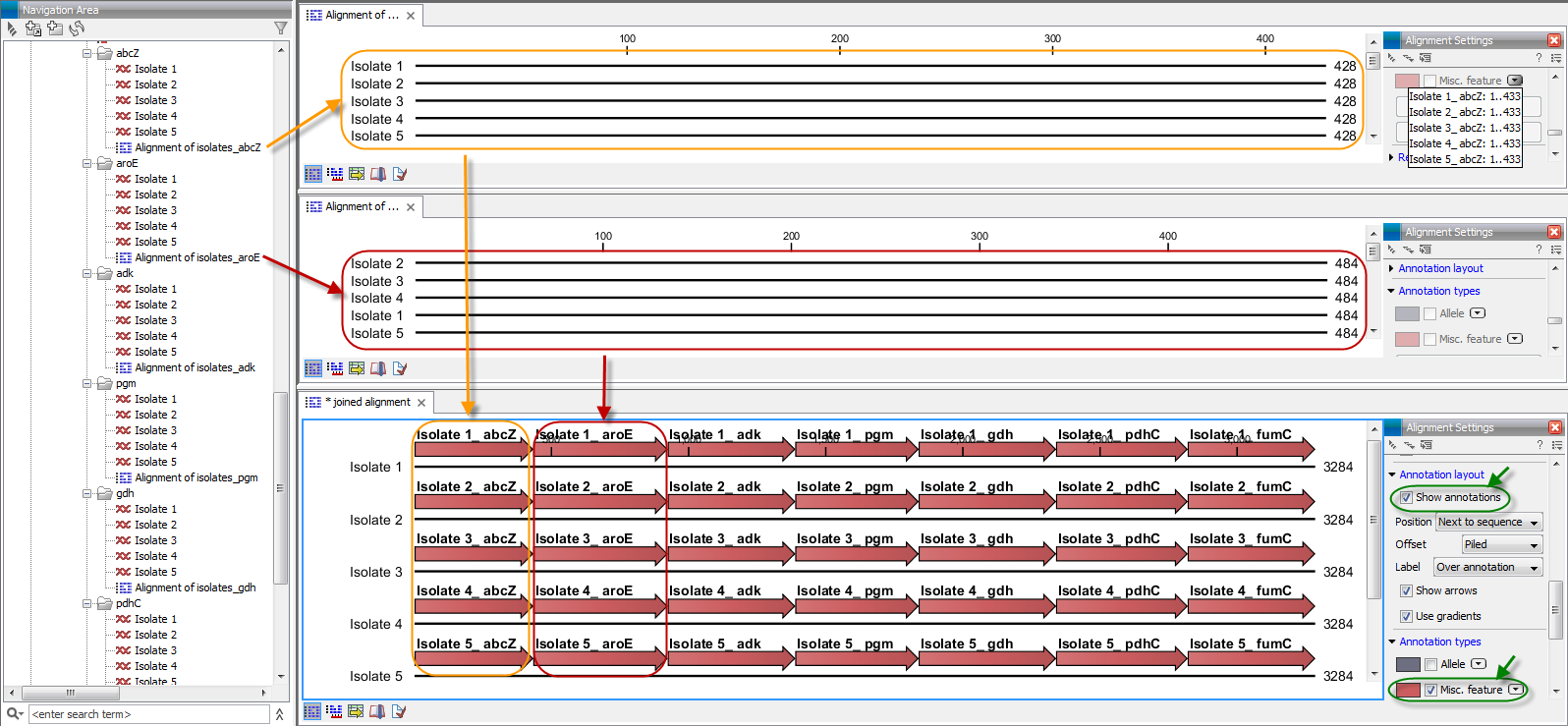
Figure 20.15: The upper part of the figure shows two of the seven alignments for the genes "abcZ" and "aroE" respectively. Each alignment consists of sequences from one gene from five different isolates. The lower part of the figure shows the result of "Join Alignments". Seven genes have been joined to an artificial gene fusion, which can be useful for construction of phylogenetic trees in cases where only fractions of a genome is available. Joining of the alignments results in one row for each isolate consisting of seven fused genes. Each fused gene sequence corresponds to the number of uniquely named sequences in the joined alignments.
How alignments are joined
Alignments are joined by considering the sequence names in the individual alignments. If two sequences from different alignments have identical names, they are considered to have the same origin and are thus joined. Consider the joining of the alignments shown in figure 20.15 "Alignment of isolates_abcZ", "Alignment of isolates_aroE", "Alignment of isolates_adk" etc. If a sequence with the same name is found in the different alignments (in this case the name of the isolates: Isolate 1, Isolate 2, Isolate 3, Isolate 4, and Isolate 5), a joined alignment will exist for each sequence name. In the joined alignment the selected alignments will be fused with each other in the order they were selected (in this case the seven different genes from the five bacterial isolates). Note that annotations have been added to each individual sequence before aligning the isolates for one gene at the time in order to make it clear which sequences were fused to each other.
