Fixpoints
With fixpoints, you can get
full control over the alignment algorithm. The fixpoints are points
on the sequences that are forced to align to each other.
To add a fixpoint, open the sequence or alignment and:
Select the region you want to use as a fixpoint | right-click the selection | Set alignment fixpoint here
This will add an annotation labeled "Fixpoint" to the sequence (see figure 20.6). Use this procedure to add fixpoints to the other sequence(s) that should be forced to align to each other.

Figure 20.6: Adding a fixpoint to a sequence in an existing alignment. At the top you can see a fixpoint that has already been added.
When you click "Create alignment" and go to Step 2, check Use fixpoints in order to force the alignment algorithm to align the fixpoints in the selected sequences to each other.
In figure 20.7 the result of an alignment using fixpoints is illustrated.
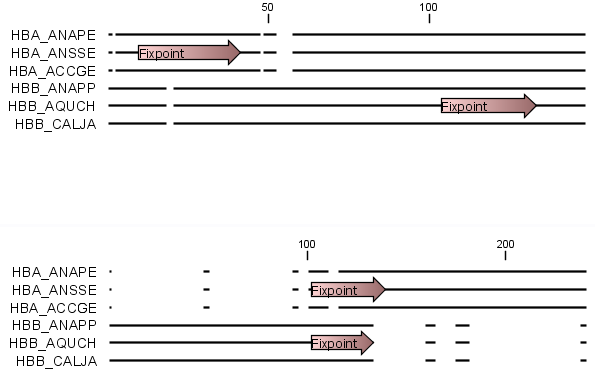
Figure 20.7: Realigning using fixpoints. In the top view, fixpoints have been added to two of the sequences.
In the view below, the alignment has been realigned using the fixpoints. The three top sequences are very similar,
and therefore they follow the one sequence (number two from the top) that has a fixpoint.
You can add multiple fixpoints, e.g. adding two fixpoints to the sequences that are aligned will force their first fixpoints to be aligned to each other, and their second fixpoints will also be aligned to each other.
Advanced use of fixpoints Fixpoints with the same names will be aligned to each other, which gives the opportunity for great control over the alignment process. It is only necessary to change any fixpoint names in very special cases.
One example would be three sequences A, B and C where sequences A and B has one copy of a domain while sequence C has two copies of the domain. You can now force sequence A to align to the first copy and sequence B to align to the second copy of the domains in sequence C. This is done by inserting fixpoints in sequence C for each domain, and naming them 'fp1' and 'fp2' (for example). Now, you can insert a fixpoint in each of sequences A and B, naming them 'fp1' and 'fp2', respectively. Now, when aligning the three sequences using fixpoints, sequence A will align to the first copy of the domain in sequence C, while sequence B would align to the second copy of the domain in sequence C.
You can name fixpoints by:
right-click the Fixpoint annotation | Edit
Annotation (![]() ) | type the name in the
'Name' field
) | type the name in the
'Name' field
