Alignment-based TaqMan probe design
CLC Genomics Workbench allows the user to design solutions for TaqMan quantitative PCR which consist of four oligos: a general primer pair which will amplify all sequences in the alignment, a specific TaqMan probe which will match the group of included sequences but not match the excluded sequences and a specific TaqMan probe which will match the group of excluded sequences but not match the included sequences. As above, the selection boxes are used to indicate the status of a sequence, if the box is checked the sequence belongs to the included sequences, if not, it belongs to the excluded sequences. We use the terms included and excluded here to be consistent with the section above although a probe solution is presented for both groups. In TaqMan mode, primers are not allowed degeneracy or mismatches to any template sequence in the alignment, variation is only allowed/required in the TaqMan probes.Pushing the Calculate button will cause the dialog shown in figure 17.15 to appear.
The top part of this dialog is identical to the Standard PCR dialog for designing primer pairs described above.
The central part of the dialog contains parameters to define the specificity of TaqMan probes. Two parameters can be set:
- Minimum number of mismatches - the minimum total number of mismatches that must exist between a specific TaqMan probe and all sequences which belong to the group not recognized by the probe.
- Minimum number of mismatches in central part - the minimum number of mismatches in the central part of the oligo that must exist between a specific TaqMan probe and all sequences which belong to the group not recognized by the probe.
The lower part of the dialog contains parameters pertaining to primer pairs and the comparison between the outer oligos(primers) and the inner oligos (TaqMan probes). Here, five options can be set:
- Maximum percentage point difference in G/C content (described above under Standard PCR).
- Maximal difference in melting temperature of primers in a pair - the number of degrees Celsius that primers in the primer pair are all allowed to differ.
- Maximum pair annealing score - the maximum number of hydrogen bonds allowed between the forward and the reverse primer in an oligo pair. This criteria is applied to all possible combinations of primers and probes.
- Minimum difference in the melting temperature of primer (outer) and TaqMan probe (inner) oligos - all comparisons between the melting temperature of primers and probes must be at least this different, otherwise the solution set is excluded.
- Desired temperature difference in melting temperature between
outer (primers) and inner (TaqMan) oligos - the scoring function discounts
solution sets which deviate greatly from this value. Regarding this,
and the minimum difference option mentioned above, please note that
to ensure flexibility there is no directionality indicated when
setting parameters for melting temperature differences between
probes and primers, i.e. it is not specified whether the probes should have a lower or higher
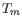 . Instead this is determined by the allowed temperature
intervals for inner and outer oligos that are set in the primer
parameters preference group in the side panel. If a higher
. Instead this is determined by the allowed temperature
intervals for inner and outer oligos that are set in the primer
parameters preference group in the side panel. If a higher 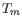 of probes is required, choose a
of probes is required, choose a 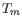 interval for probes which has higher
values than the interval for outer primers.
interval for probes which has higher
values than the interval for outer primers.
The output of the design process is a table of solution sets. Each solution set contains the following: a set of primers which are general to all sequences in the alignment, a TaqMan probe which is specific to the set of included sequences (sequences where selection boxes are checked) and a TaqMan probe which is specific to the set of excluded sequences (marked by *). Otherwise, the table is similar to that described above for TaqMan probe prediction on single sequences.
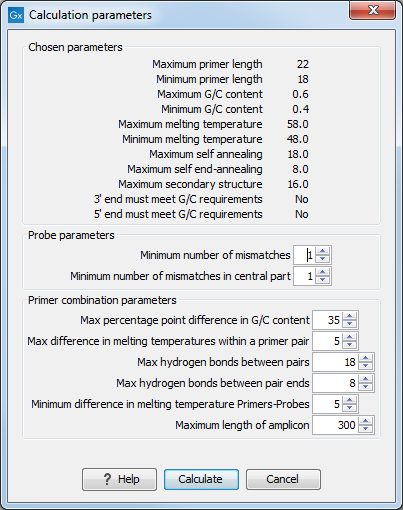
Figure 17.15: Calculation dialog shown when designing alignment based TaqMan probes.
