Running tools
All the analyses in the Toolbox are performed in a step-by-step
procedure:
- Data elements to be used in the analysis are selected.
- Any configurations necessary for the tool to run are made.
- The results are opened or saved.
You can open a tool from the Toolbox by double clicking on its name in the Toolbox in the bottom left side of the Workbench, or by selecting it from the Toolbox menu at the top. You can also find tools quickly by clicking on the Launch button (![]() ) in the toolboar. Double click on the name of the tool in the table to launch it. If you enter a search term in the field at the top of the Quick launch window, tools with that term in their names or descriptions will be listed.
) in the toolboar. Double click on the name of the tool in the table to launch it. If you enter a search term in the field at the top of the Quick launch window, tools with that term in their names or descriptions will be listed.
When you open a tool, a wizard pops up in the center of the View Area. Stepping through a succession of wizard steps, you will select the data to analyze, configure any analysis parameters, and specify how the results should be handled. You can navigate between wizard steps by clicking the buttons Next and Previous at the bottom of the window.
If you have logged into a CLC Server from your Workbench, you will first be asked to select whether the job should be run on the Workbench or submitted to the server. These choices, along with information about data selection and other considerations when launching tasks on a CLC Server are provided in Running a tool on a CLC Server.
Generally, the first analysis configuration step involves selecting the data elements to be used as input. A view of your Navigation Area will be presented to you. That view will show data elements appropriate for use as input for that tool. Folders are also shown. For example, in figure 9.1 you can see a the Workbench Navigation Area (on the left) and a view of the same Navigation Area in the wizard (on top) for the Assemble Sequences tool. This tool only accepts nucleotide sequences and nucleotide sequence lists, so data elements of other types that can be seen in the Workbench Navigation Area, such as the one called "Read mapping", and the amino acid sequence ATP8a1, are not displayed in the wizard Navigation Area.
The data types that can be used as input for a given tool are described in the manual section about that tool. This documentation can be opened directly by clicking on the Help button in the bottom left corner of the launch wizard.
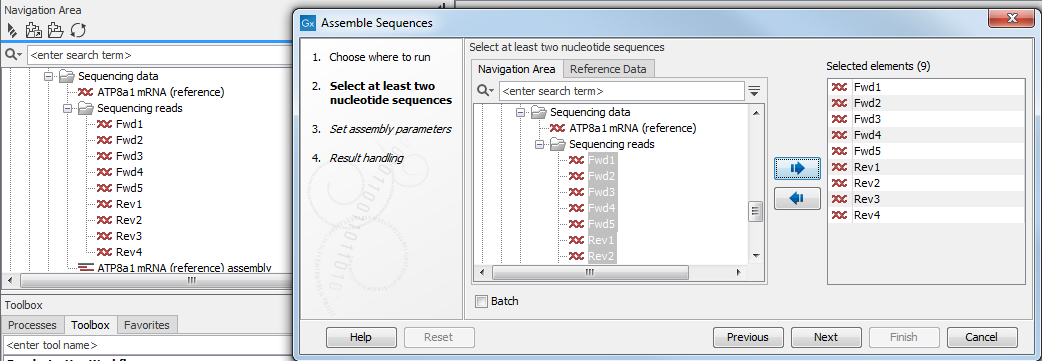
Figure 9.1: You can select input files for the tool from the Navigation Area view presented on the left hand side of the wizard window.
To indicate the data elements to be used in the analysis, either double click on them in the "Navigation Area" view on the left, or select them with a single click and then click on the right hand arrow. These items will then be listed in the "Selected elements" list on the right. If data elements of appropriate types were already selected in the Workbench Navigation Area before launching the tool, these will be automatically entered into the Selected elements list. To remove entries in that list, just double click on them or select them with a single click and then click on the left hand arrow.
When multiple elements are selected, most analysis tools will treat all those elements as a single input data set unless the "Batch" option at the bottom, has been selected. If that option is selected, then the tool will be run multiple times, once for each "batch unit", which may be a data element, or folder containing data elements or containing folders of elements. Batch processing is described in more detail in Batch processing.
Once the data of interest has been selected, click on Next. Depending on the tool, there may now be one or more steps for configuring analysis parameters. An example is shown in figure 9.2. Clicking on the Reset button resets all parameters in that step to their default values.
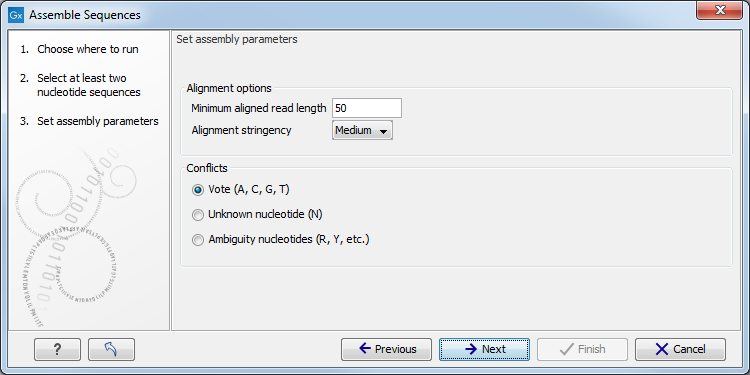
Figure 9.2: An example of a "Set parameters" window.
