De novo assembly output
Stand-alone mapping. The de novo assembly is followed by a step where the reads used to generate the contigs are mapped against the contigs, using them as reference.
This output is called assembly and opens as a table listing all contigs as seen in figure 30.22.
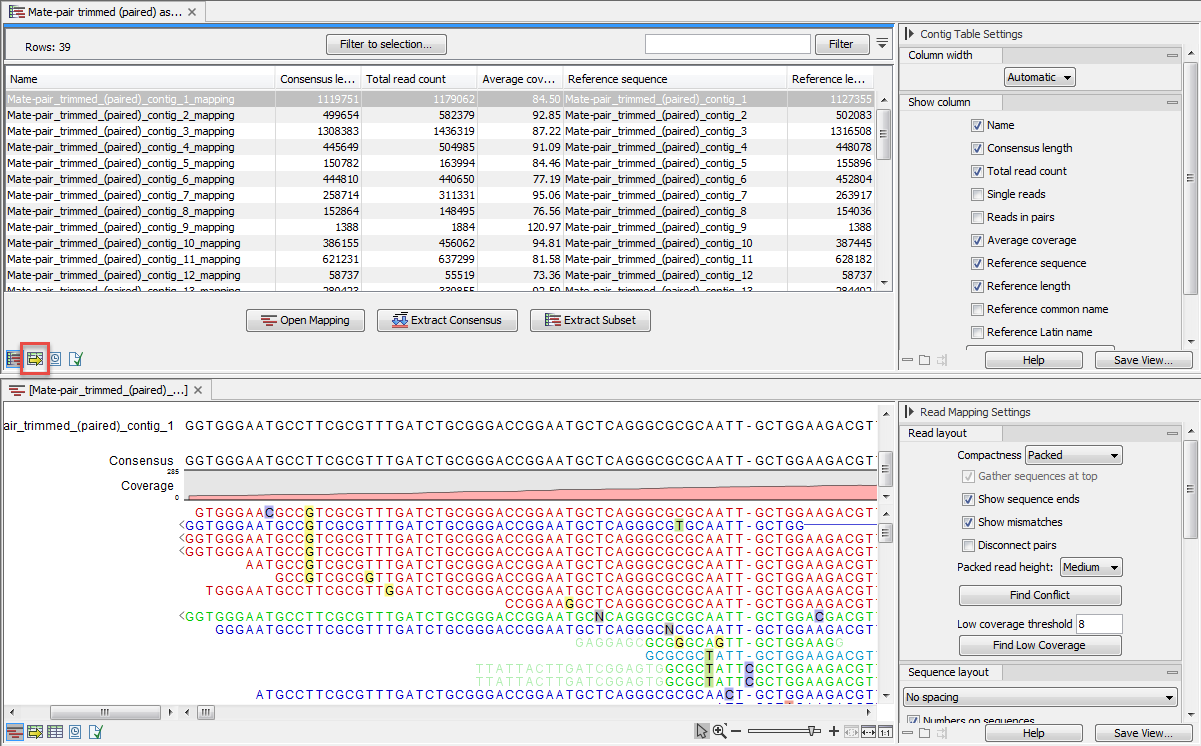
Figure 30.22: A de novo assembly read mapping.
The information included in the table is:
- Name. When mapping reads to a reference, this will be the name of the reference sequence.
- Consensus length. The length of the consensus sequence. Subtracting this from the length of the reference will indicate how much of the reference that has not been covered by reads.
- Total read count. The number of reads. Reads with multiple hits on different reference sequences are placed according to your input for Non-specific matches
- Single reads and Reads in pair. Total number of reads, single and/or in pair.
- Average coverage. This is simply summing up the bases of the aligned part of all the reads divided by the length of the reference sequence.
- Reference sequence. The name of the reference sequence.
- Reference length. The length of the reference sequence.
- Reference common name and Reference latin name. Name, common name and Latin name of each reference sequence.
At the bottom of the table there are three buttons that can be used to open or extract sequences. Select the relevant rows before clicking on the buttons:
- Open Mapping. Opens the read mapping for visual inspection. You can also open one mapping simply by double-clicking in the table.
- Extract Consensus/Contigs. For de novo assembly results, the contig sequences will be extracted. For results when mapping against a reference, the Extract Consensus tool will be used (see Extract consensus sequence).
- Extract Subset. Creates a new mapping table with the mappings that you have selected.
Double clicking on a contig name will open the read mapping in split view.
It is possible to open the assembly as an annotation table (using the icon highlighted in figure 30.22). The annotations available in the table are the following (see figure 30.23):
- Alternatives Excluded. More than one path through the graph was possible in this region but evidence from paired data suggested the exclusion of one or more alternative routes in favour of the route chosen.
- Contigs Joined. More than one route was possible through the graph such that an unambiguous choice of how to traverse the graph cannot by made. However evidence from paired data supports one of these routes and on this basis, this route is selected (and other routes excluded).
- Scaffold. The route through the graph is not clear but evidence from paired data supports the connection of two contigs. A single contig is then reported with N characters between the two connected regions. This entity is also known as a scaffold. The number of N characters represents the expected distance between the regions, based on the evidence the paired data.
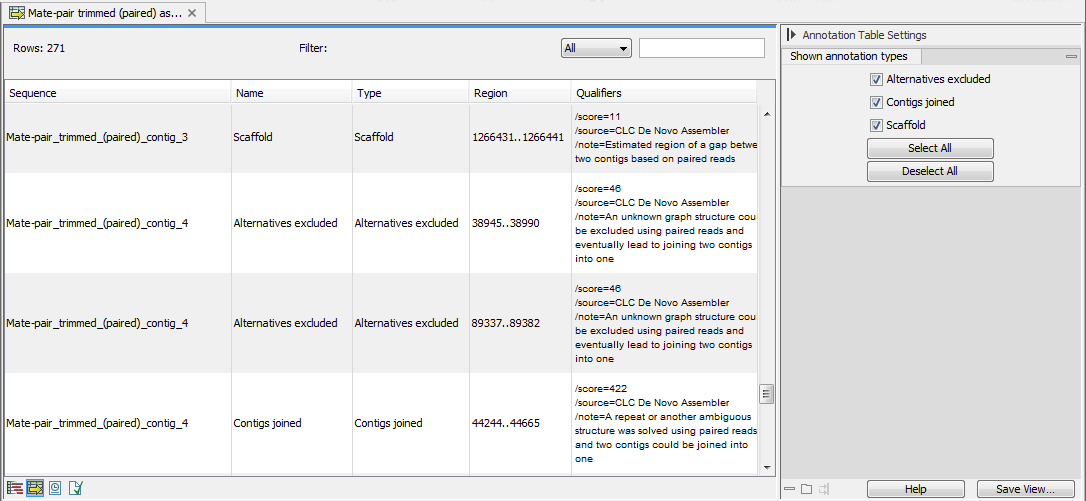
Figure 30.23: A de novo assembly read mapping seen as annotation table.
Using the menu in the right end side panel, it is possible to select only one type of annotations to be displayed in the table.
Simple contigs. The output is a sequence list of the contigs generated (figure 30.24), that can also be seen as a table and an annotation table as described for the stand-alone read mapping described above.
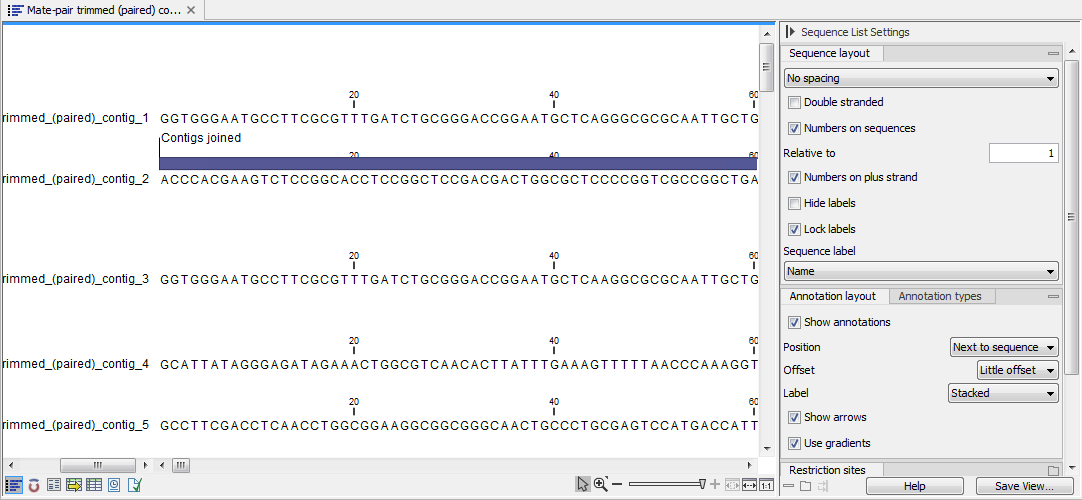
Figure 30.24: A de novo assembly read mapping seen as annotation table.
