Specific options for alignment-based primer and probe design
Compared to the primer view of a single sequence, the most notable difference is that the alignment primer view has no available graphical information. Furthermore, the selection boxes found to the left of the names in the alignment play an important role in specifying the oligo design process.The Primer Parameters group in the Side Panel has the same options as the ones defined for primers design based on single sequences, but differs by the following submenus(see figure 17.12):
- In the Mode submenu, specify either:
- Standard PCR. Used when the objective is to design primers, or primer pairs, for PCR amplification of a single DNA fragment.
- TaqMan. Used when the objective is to design a primer pair and a probe set for TaqMan quantitative PCR.
- In the Primer solution submenu, specify requirements for the match of a PCR primer against the template sequences.
These options are described further in Alignment based design of PCR primers. It contains the following options:
- Perfect match
- Allow degeneracy
- Allow mismatches
The workflow when designing alignment based primers and probes is as follows (see figure 17.12):
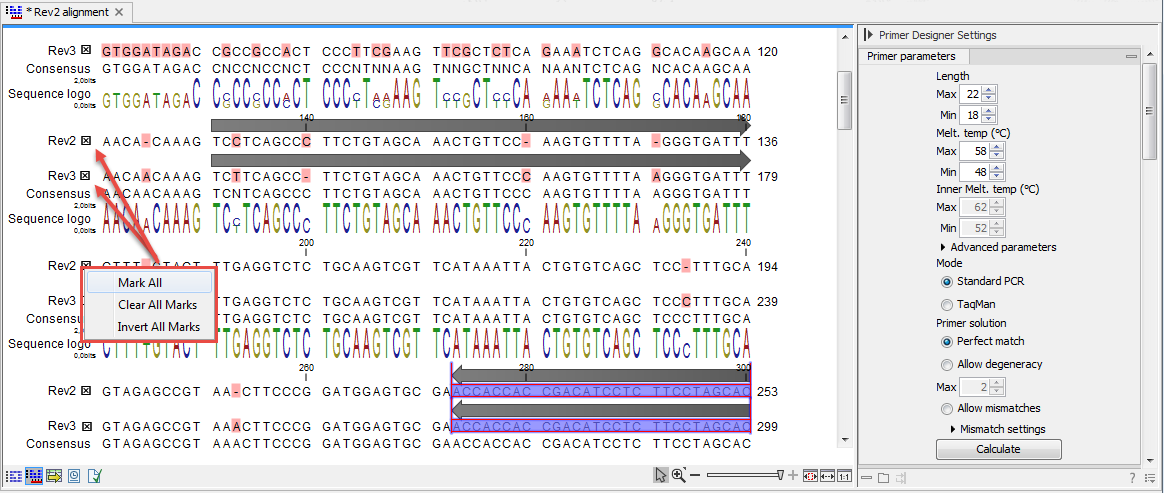
Figure 17.13: The initial view of an alignment used for primer design.
- Use selection boxes to specify groups of included and excluded sequences. To select all the sequences in the alignment, right-click one of the selection boxes and choose Mark All.
- Mark either a single forward primer region, a single reverse primer region or both on the sequence (and perhaps also a TaqMan region). Selections must cover all sequences in the included group. You can also specify that there should be no primers in a region (No Primers Here) or that a whole region should be amplified (Region to Amplify).
- Adjust parameters regarding single primers in the preference panel.
- Click the Calculate button.
