Type With Large MLST Scheme
The Type With Large MLST Scheme tool is used for assigning a sequence type to an isolate.
To run the Type With Large MLST Scheme tool choose:
Microbial Genomics Module (![]() ) | Typing and Epidemiology (
) | Typing and Epidemiology (![]() ) | Large MLST Typing (
) | Large MLST Typing (![]() ) | Type With Large MLST Scheme (
) | Type With Large MLST Scheme (![]() )
)
The tool takes a sequence list as input and will work with either raw NGS reads or an assembled genome. Note that if the input is raw NGS reads, and the tool reports multiple ambiguous sequence types, performing a standard De Novo Assembly might help to reduce noise and provide a more conclusive typing result.
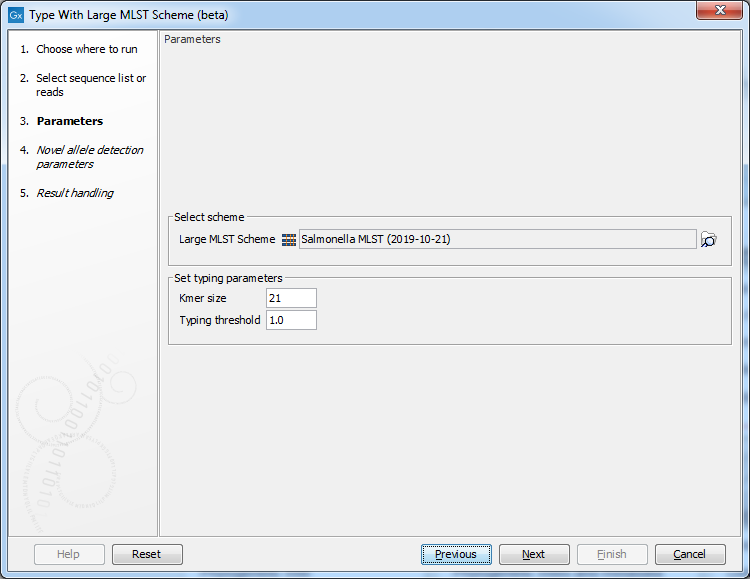
Figure 13.12: Specifying scheme and typing parameters.
In the next dialog step (figure 13.12), specify the scheme and the typing parameters.
The tool works by comparing the kmers in the input to the kmers in the alleles for the different loci.
The Kmer size determines the number of nucleotides in the kmer - raising this setting might increase specificity at the cost of some sensitivity.
The Typing threshold determines how many of the kmers in a sequence type that needs to be identified before a typing is considered conclusive. The default setting of 1.0 means that all kmers in all alleles must be matched. Lowering the setting to 0.99 would mean that on average 99% of the kmers in all the alleles of a given sequence type must be detected before the sequence type is considered conclusive.
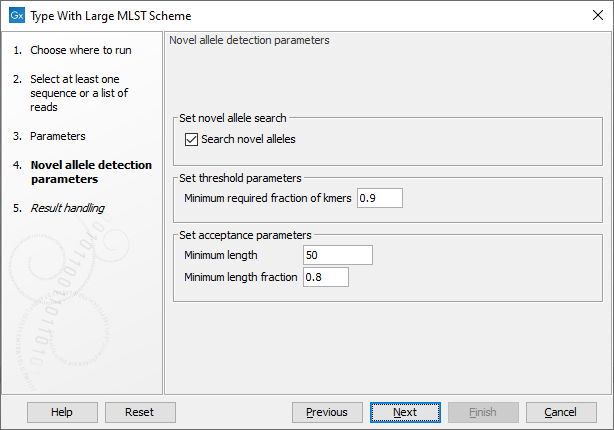
Figure 13.13: Specifying novel allele detection parameters.
The next step in the dialog determines how to handle novel alleles (figure 13.13): if the input isolate has loci with alleles that are not part of the scheme, it is possible to still detect the novel alleles. The novel alleles and the resulting new sequence type can then be added to the scheme using the Add Typing Results to Large MLST Scheme tool.
Novel alleles are detected as close hits to existing alleles in a locus. The Minimum required fraction of kmers determines how close a match must be: the default setting of 0.9 means that at least 90% of the kmers for an allele in a locus must be identified before the novel allele detection is initiated.
If the input to the tool is raw NGS reads, the tool will assemble the reads containing the kmers for the possible novel allele. If the input is already an assembled genome, the existing alleles for a locus will be mapped to the assembly to extract a novel allele.
After a candidate novel allele has been identified, it is aligned to the other alleles in the locus.
If the scheme has been built with the Check codon positions option of the Create Large MLST Scheme tool enabled (see Create Large MLST Scheme)), or if the scheme was imported with a specified genetic code (see Download Large MLST Scheme), the start and stop codons in the novel allele sequence are then identified, and the sequence is then trimmed to the start and stop codons that most closely match the length of the existing alleles in the locus. Alleles that contain both a start and a stop codon at the beginning and end, respectively, and pass the acceptance parameters (see below) will be marked as Complete in the output table from the tool.
The acceptance parameters describe the final consistency check: the novel allele must not contain a stop codon, it must be at least the Minimum length in nucleotides and have at least a length of the specified Minimum length fraction of the shortest allele in the locus before it is accepted.
Subsections
