Map to Specified Reference
Once analysis has been performed using the Type Among Multiple Species workflow, the best matching reference is listed in the Result Metadata table (figure 10.1, see column Best match).
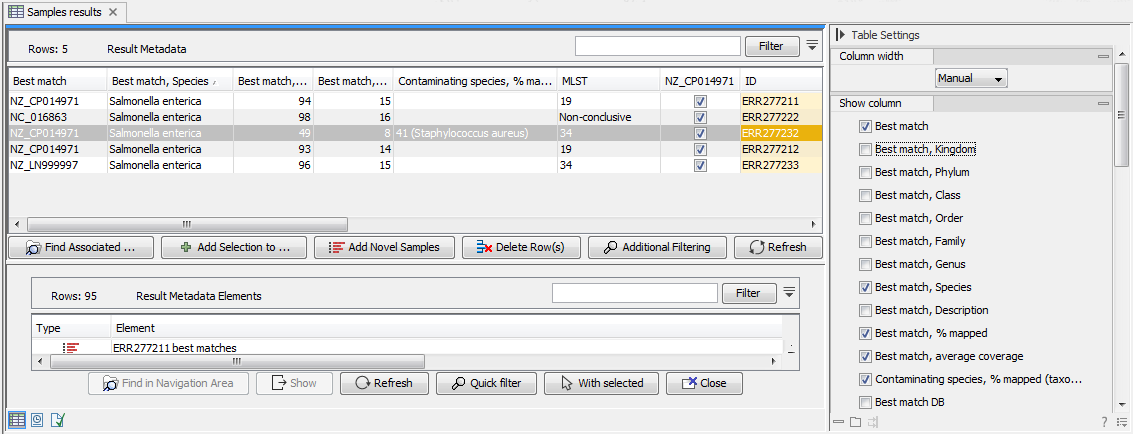
Figure 10.1: Best match references are listed for each row in the Result Metadata Table.
If all your samples share the same common reference, you can proceed to additional analyses without delay.
However there are cases where your samples have different Best match reference for a particular MLST scheme. And because creating a SNP Tree require a single common reference, you will need to identify the best matching common reference for all your samples using a K-mer Tree, as well as subsequently re-map your samples to this common reference.
If you already know the common reference for the sample you want to use to create a SNP tree, you can directly specify that reference in the re-map workflow. Otherwise, finding a common reference is described in more details in section 11.2.
In short, to identify a common reference across multiple clades within the Result Metadata Table:
- Select samples to which a common best matching references should be identified.
- Click on the Find Associated Data (
 ) button to find their associated Metadata Elements.
) button to find their associated Metadata Elements.
- Click on the Quick Filtering (
 ) button and select the option Filter for K-mer Tree to find Metadata Elements with the Role = Trimmed Reads.
) button and select the option Filter for K-mer Tree to find Metadata Elements with the Role = Trimmed Reads.
- Select the relevant Metadata Element files.
- Click on the With selected (
 ) button.
) button.
- Select the Create K-mer Tree action and follow the wizard as described in the section called Create K-mer Tree.
The common reference, chosen as sharing the closest common ancestor with the clade of isolates under study in the k-mer tree, is subsequently used as a reference for the Map to Specified Reference workflow (figure 10.2) that will perform a re-mapping of the reads followed by variant calling.
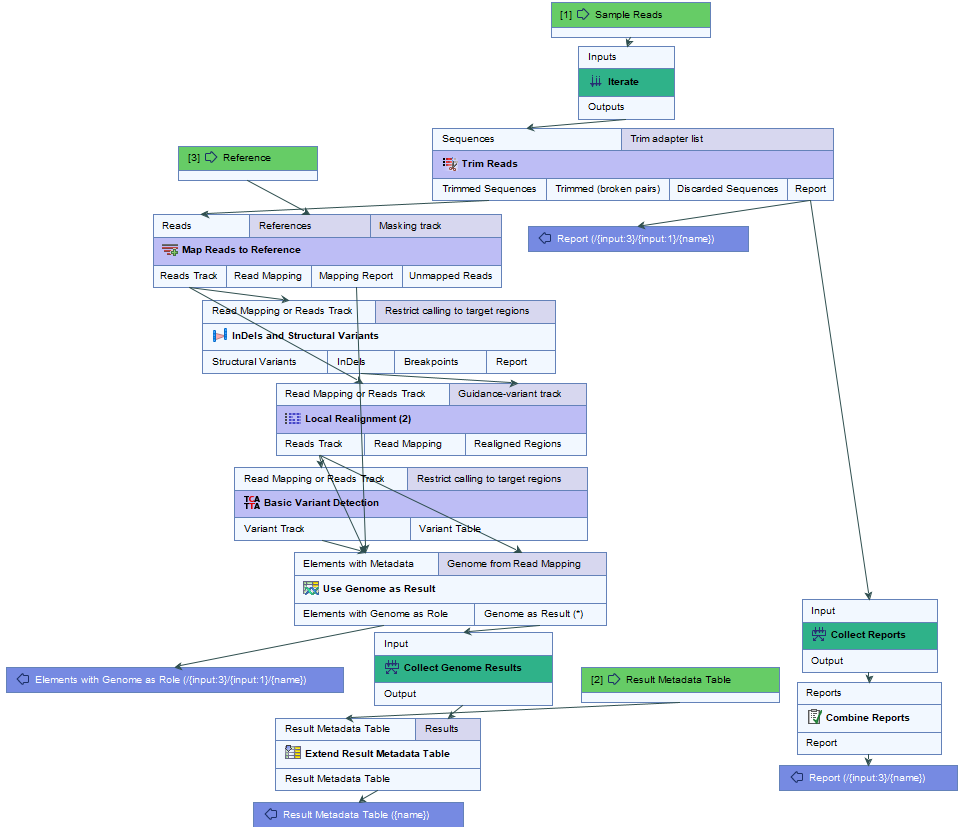
Figure 10.2: Overview of the template Map to Specified Reference workflow.
Subsections
