Estimate Alpha and Beta Diversities workflow
The Estimate Alpha and Beta Diversities workflow consists of 5 tools and requires only the OTU table as input file (figure 5.13).
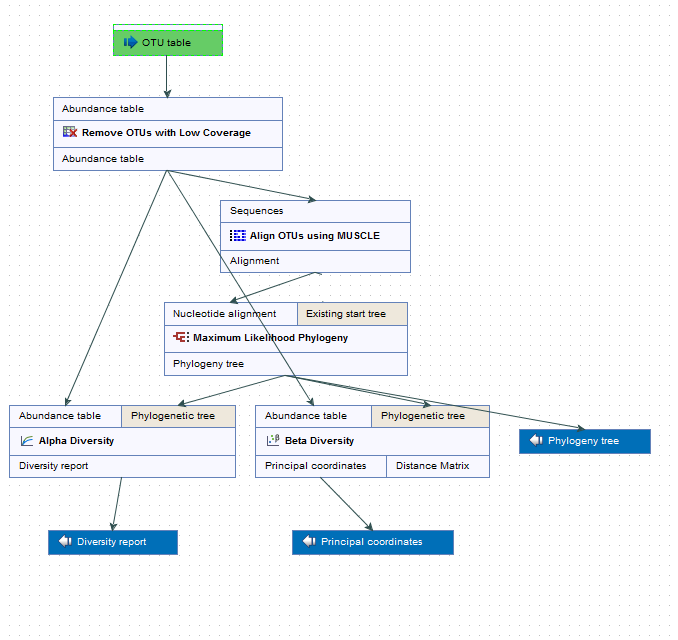
Figure 5.13: Layout of the Alpha and Beta Diversities workflow.
Remember to add metadata to the abundance table before starting the workflow. Adding metadata can be done very early on, by importing metadata and associating reads to it before generating an OTU abundance table with the OTU Clustering tool or the Data QC and OTU Clustering workflow. The metadata will propagate to the abundance table automatically. When working with reads that were not associated with metadata in the first place, it is always possible to add metadata to an already existing abundance table with the tool Add Metadata to Abundance Table.
The first tool of the workflow is the Filter OTUs Based on the Number of Reads. The output is a reduced abundance table that will be used as input for three other tools:
- Align OTUs with MUSCLE, a tool that will produce an alignment used to reconstruct a Maximum Likelihood Phylogeny (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Maximum_Likelihood_Phylogeny.html), which will in turn output a phylogenetic tree also used as input in the following two tools.
- Alpha diversity tool
- Beta diversity tool
Running this workflow will therefore give the following outputs: a phylogenetic tree of the OTUs, a diversity report for the alpha diversity and a PCoA for the beta diversity.
