Sorted Reads Files
The taxonomic profiling tool can sort reads based on their assigned position in the databases (figure 6.14): database matches, host matches, and unclassified reads.
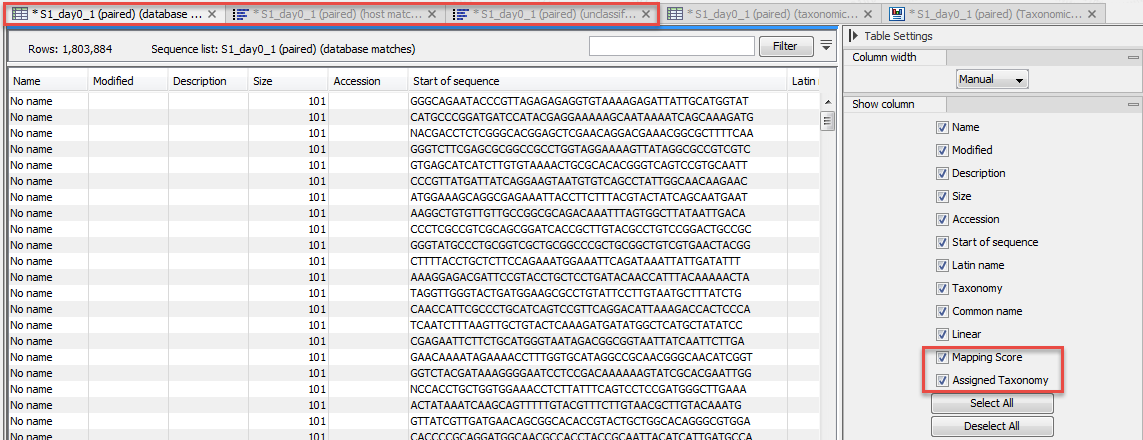
Figure 6.14:
Database matches table.
The resulting files are sequence lists that can be visualized as table. The host and database matches tables include a value called Mapping score, and calculated for each read at their assigned positions (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Mapping_parameters.html). In addition, the database matches table provide the Assigned taxonomy for the read, taxonomy that was assigned to the read by mapping it to the database.
If the reads matching the reference database have been saved, it is possible to select a set of rows in the abundance table and to click the "Extract Reads from Selection" button which will extract reads uniquely associated to the specific rows selected in the abundance table.
