Annotate CDS with Best BLAST Hit
The Annotate CDS with Best BLAST Hit tool will allow you to annotate a set of contigs containing CDS annotations with their best BLAST hit.
To start the analysis, go to:
Functional Analysis (![]() ) | Annotate CDS with Best BLAST Hit (
) | Annotate CDS with Best BLAST Hit (![]() )
)
Several parameters are available:
- Genetic code. The genetic code used for translating CDS to proteins.
- BLAST database. A protein BLAST database. Popular BLAST protein databases can be downloaded using the Download BLAST Database tool or created using a the Create BLAST Database tool.
- Maximum E-value. Maximum expectation value (E-value) threshold for saving hits.
Metadata from the sequences used to create the BLAST database (such as GO terms or taxonomy information) will not be transferred by this tool. If metadata is relevant, consider using the Annotate CDS with Best DIAMOND Hit tool instead.
Note that choosing a very large BLAST database with millions of sequences (e.g. the nt, nr and refeseq_protein databases from the NCBI) will slow down the algorithm considerably, especially when there are many CDS in the input. Therefore, we recommend to use a medium-sized database such as "swissprot".
In the wizard, you can choose between databases stored locally (![]() ) or remotely on the server (
) or remotely on the server (![]() ). If you create a workflow that you plan to run on a server, you should avoid locking the BLAST database parameter as the chosen database may not exist on the server.
). If you create a workflow that you plan to run on a server, you should avoid locking the BLAST database parameter as the chosen database may not exist on the server.
If you select Create Report, the tool will create a summary report table. The report is divided in three parts:
- Input. Contains information about the size of the contigs and CDS used as input.
- BLAST database. The protein BLAST database used in the search, together with its description, location, and size.
- Output. The total number (and percent) of CDS that were annotated with their best BLAST hit.
The tool will output a copy of the input file containing the following fields when a hit for a CDS is found (figure 14.8):
- BLAST Hit. Accession number of the best BLAST Hit in the BLAST database.
- BLAST Hit Description. Description of the matching protein, as present in the BLAST database.
- BLAST Hit E-value. The E-value of the match.
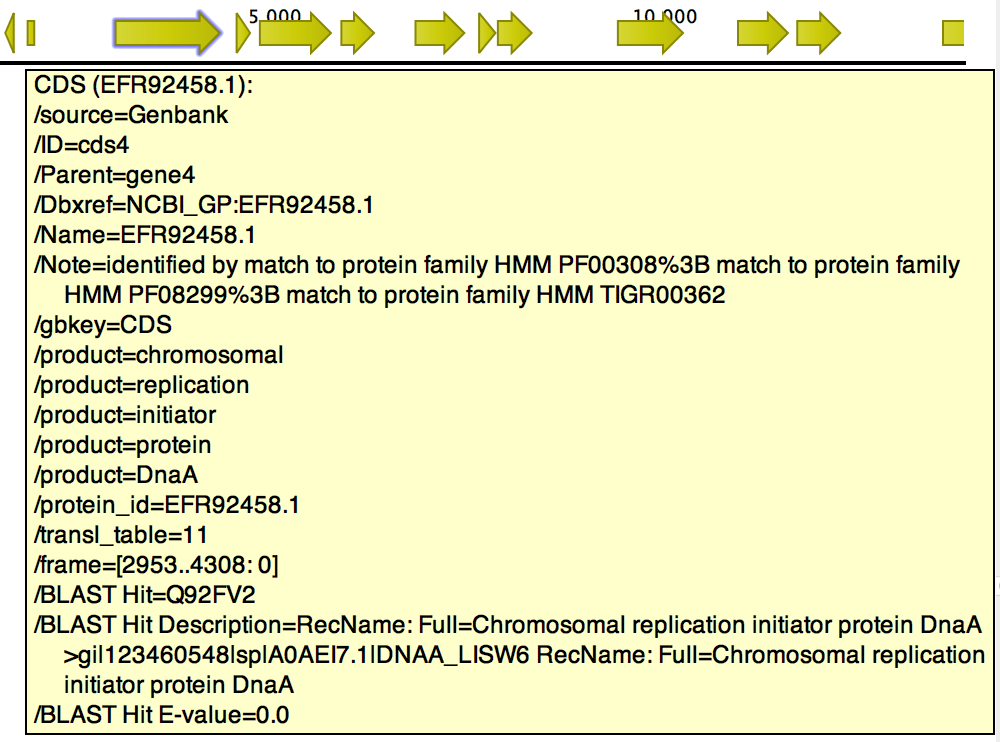
Figure 14.8: BLAST Best Hit annotations added to gene cds4 of h. pylori.
The tool can also output an annotation table summarizing information about the annotations added to the sequence list.
