Download Custom Microbial Reference Database
The Download Custom Microbial Reference Database tool allows you to create a custom database from taxonomies or NCBI assembly IDs. The tool outputs a single sequence list. It is possible to download the sequence list with the necessary annotations required for the tools in the Typing and Epidemiology and Metagenomics sections of the Microbial Genomics Module.
To run the tool, go to:
Databases (![]() ) | Taxonomic Analyses (
) | Taxonomic Analyses (![]() ) | Download Custom Microbial Reference Database (
) | Download Custom Microbial Reference Database (![]() )
)
In the first window (figure 18.3) in the section Customize Database, select one of the three options:
- Include all: The database will contain both genomic and plasmid sequences.
- Include only plasmids: The database will contain only plasmid sequences.
- Exclude all plasmids: The database will not contain any plasmid sequences.
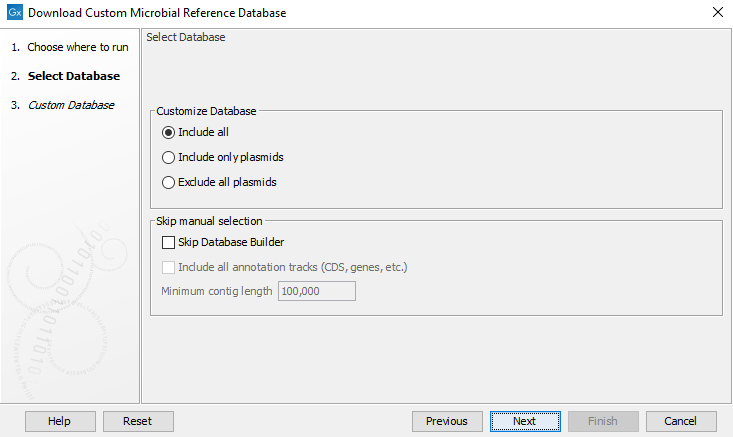
Figure 18.3: Select the source of the database
Here you can also choose to skip the database builder. If you choose to skip, all references matching the criteria stated in the next step will be downloaded. Selecting skip database builder will also enable the options to include all annotation tracks (annotation tracks are not needed for taxonomic profiling applications, but required when creating Large MLST schemes). An additional filter, Minimum contig length, may also be specified. Note that if you have not selected to skip the database builder, these options can be set later when downloading the database.
Click Next to customise the database you wish to generate (as in figure 18.4). You have the following two options:
- Build database from accessions or species TaxIDs: This enables the ID matching field. Here you can provide a list of Species TaxIDs or GenBank or RefSeq assembly accessions (one per line) that must be included in your database. If using GenBank or RefSeq assembly accessions, the accessions must follow the assembly accession: 3 letter prefix, (GCA for GenBank assemblies or GCF for RefSeq assemblies) followed by an underscore and 9 digits. For example, GCA_000019425 for the assembly of the DH10B substrain of E. coli. If a version number is included, it will be ignored and the newest version downloaded.
The GenBank assembly accession can usually be accessed using the search function or from the GenBank sequence on NCBI.
Species TaxIDs, GenBank or RefSeq assembly accessions can also be provided in a .txt file by clicking Browse.
- Build database from taxonomic lineages: This enables the Linages prefixes field. Here you can provide a list of taxonomic lineage prefixes (one per line) to include in your database. The lineages should follow the format of 7-step taxonomies. For example entering "Bacteria;Firmicutes;Bacilli;Staphylococcales;Staphylococcaceae;" will include all genera and species references in the Staphylococcaceae family. Entering "Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;" will include all family, genera and species references in the Enterobacterales order.
Lineage prefixes can also be provided in a .txt file by clicking Browse.
The size and completeness of the database can be tuned with Inclusion criteria which select representative references based on the following options:
- All reference genomes: all reference genomes in the chosen lineage(s) are included.
- All representative genomes: all representative genomes in the chosen lineage(s) are included.
- All reference and representative genomes: all reference and representative genomes in the chosen lineage(s) are included.
- All genomes: all genomes in the chosen lineage(s) are included.
- One reference per species: One reference is selected for each species in the chosen lineage(s). The chosen species representative is selected based on ranking with Reference genomes > Representative genomes > Complete genomes > Scaffolds > Contigs. When two or more references share the same rank, the reference with the longest chromosome is selected. Note species are identified using species TaxIDs. This means that assemblies with different species names but the same TaxIDs are considered as one species.
- One reference per genus: One reference is selected for each genus in the chosen lineage(s). The chosen genus representative is selected based on ranking with Reference genomes > Representative genomes > Complete genomes > Scaffolds > Contigs. When two or more references share the same rank, the reference with the longest chromosome is selected.
It is also possible to leave the Linages prefixes and ID matching fields empty. In this case the database will not have any references pre-selected. Selection can then be done manually in the builder.
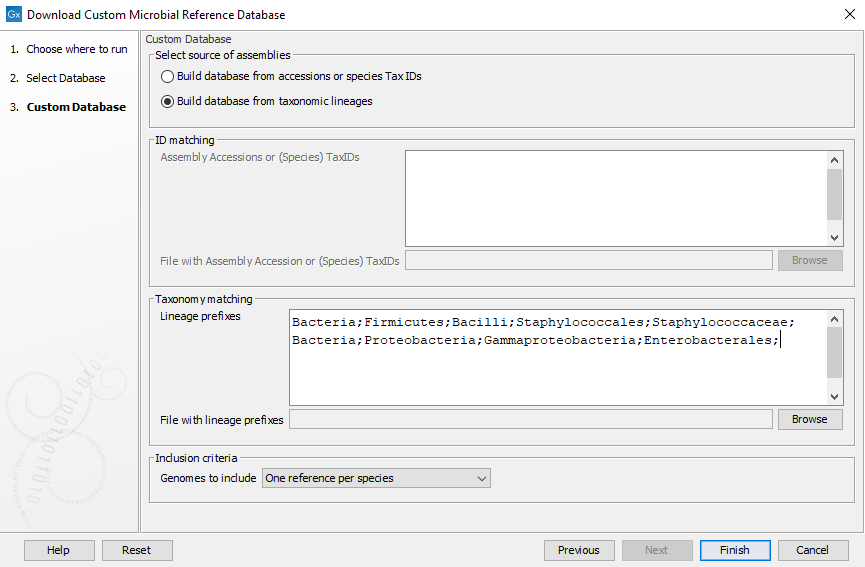
Figure 18.4: Select options for building the database
If Skip Database Builder was enabled, all references matching the specified criteria will be downloaded and the running of the tool is complete and you will not see the Database Builder as described in the next section.
Subsections
