Download Curated Microbial Reference Database
The Download Curated Microbial Reference Database tool downloads selected reference databases as single sequence lists and/or taxonomic profiling indices with the necessary annotations required for the tools in the Typing and Epidemiology and Metagenomics sections of the Microbial Genomics Module.
To run the tool, go to:
Databases (![]() ) | Taxonomic Analyses (
) | Taxonomic Analyses (![]() ) | Download Curated Microbial Reference Database (
) | Download Curated Microbial Reference Database (![]() )
)
In the first window (figure 18.1), select the database you wish to download.
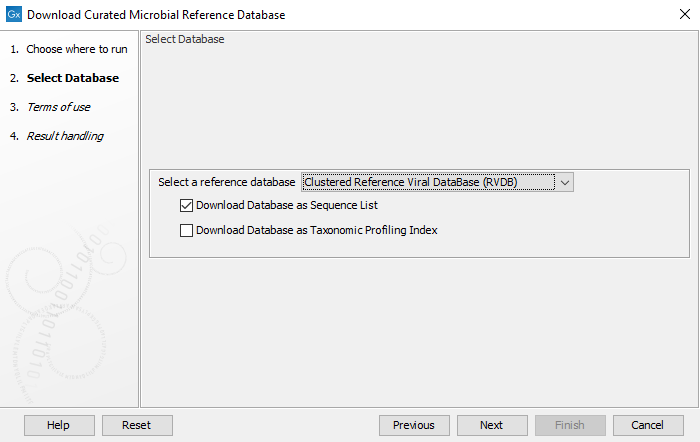
Figure 18.1: Select the database and output format
You can choose between several databases
- QMI-PTDB June 2021 - Approx. 22GB memory required: QIAGEN Microbial Insights - Prokaryotic taxonomy database is a microbial reference database for taxonomic profiling of bacteria and archaea. This database contains additional references not present in the smaller version. It is not suitable for running on a standard laptop.
- QMI-PTDB June 2021 - Approx. 16GB memory required: QIAGEN Microbial Insights - Prokaryotic taxonomy database is a microbial reference database for taxonomic profiling of bacteria and archaea. This is a subset of the larger database suitable for running on a standard laptop.
- Clustered Reference Viral DataBase (RVDB): Clustered Reference Viral Database for virus detection.
- Unclustered Reference Viral DataBase (RVDB): Unclustered Reference Viral Database for virus detection.
You can then chose to download the database as an annotated sequence list and/or as a taxonomic profiling index.
The Curated Microbial Reference Databases are optimized for balance in the taxonomic representation across the taxonomy, i.e. the oversampling of some branches of the taxonomy is removed by using representative sequences. This has the consequence that some assemblies may not be particularly good assemblies, yet they are included as they constitute the best current representative of the given branch in the taxonomy. For this optimized database you can choose to download the 22g database, or one that is optimized for running the Taxonomic Profiling tool on a laptop computer with 16GB of main memory. The 16g version of the curated database contain a smaller number of assemblies, in order to be able to run on a system with 16GB of main memory.
Note: some of the databases offered are derived works, licensed under a Creative Commons Attribution-ShareAlike (CC BY-SA) license. We offer free access to those without requiring a CLC product license: they can be downloaded using a trial version of the Microbial Genomics Module, or our support team can be contacted at ts-bioinformatics@qiagen.com to get a download link. The downloaded files can then be exported to non-proprietary formats using the freely available viewing mode of the CLC Genomics Workbench.
Subsections
