Mapping output options
Click Next lets you choose how the output of the mapping should be reported (see figure 19.10).
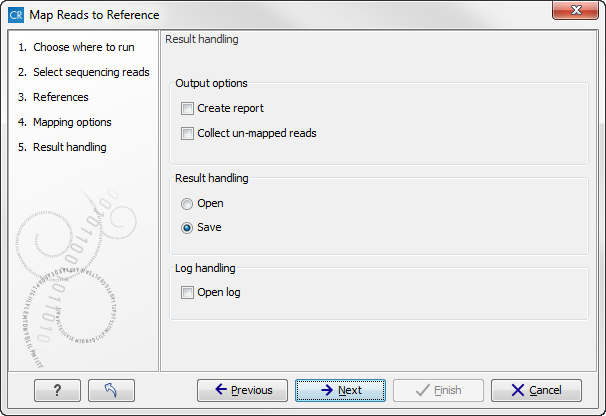
Figure 19.10: Mapping output options.
There are two independent output options available that can be (de-)activated in both cases:
- Create report. This will generate a summary report as described in Summary mapping report.
- Collect un-mapped reads. This will collect all the reads that could not be mapped to the reference into a sequence list (there will be one list of unmapped reads per sample, and for paired reads, there will be one list for intact pairs and one for single reads where the mate could be mapped).
However, the main output is a reads track:
- Reads track
- A reads track is very "lean" (i.e. with respect to memory requirements) since it only contains the reads themselves. Additional information about the reference, consensus sequence or annotations can be added and viewed alongside in the context of a Track List later (by adding, for example, a reference and/or annotation track, respectively). This kind of output is useful when working with tracks in general and especially for resequencing purposes this is recommended. Details about viewing and editing of reads-tracks are described in Genome browser.
Note that the Map Reads to Reference tool will output an empty read mapping and report when nothing mapped, and empty unmapped reads if everything mapped.
Finally, you can choose to save or open the results, and if you wish to see a log of the process, see how to handle the results.
Clicking Finish will start the mapping.
