Add information from overlapping genes
This will create a copy of the track used as input and add information from overlapping genes and mRNA tracks. To run the Add information from overlapping genes tool, go to the toolbox:
Toolbox | Add Information to Variants (![]() ) | Add Information from Overlapping Genes (
) | Add Information from Overlapping Genes (![]() )
)
First, select the track you wish to annotate and click Next. You can choose any kind of variant or annotation track as input. Next, select the a gene track and a mRNA track for overlap comparison (figure 20.12).
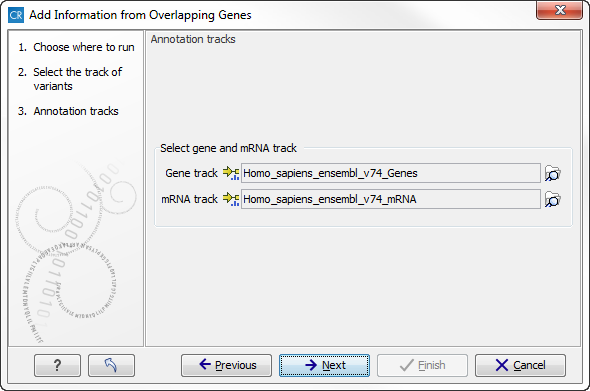
Figure 20.12: Select the genes and mRNA tracks, which can be found in the CLC_References folder.
You can find the gene and mRNA tracks in the CLC_References folder (figure 20.13).
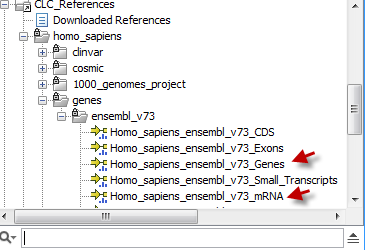
Figure 20.13: Find the genes and mRNA tracks in the CLC_References folder.
The result of this tool is a new track showing all the variants that now have been annotated with the information about genes and mRNA that overlap with the identified variants. The requirement for being registered as an overlap is that parts of the annotations are overlapping, regardless of the strandedness of the annotations (note that this makes it unsuitable for comparing e.g. two gene tracks but great for annotating variants with overlapping genes or regulatory regions). The added information can be visualized in two ways; 1) In the track tooltips when mousing over the individual variants or 2) in the table view where you can see that new columns describing the added gene and mRNA tracks have been added to the table. The table view can be accessed by clicking on the table icon (![]() ) in the lower part of the View Area.
) in the lower part of the View Area.
