Create atom group
(Often it is convenient to use a unique visualization style or color to highlight a particular set of atoms, or to visualize only a subset of atoms from a molecule. This can be achieved by creating an atom group. Atom groups can be created based on atoms selected in the 3D view or entries selected in the Project Tree. When an atom group has been created, it appears as an entry in the Project Tree in the category "Atom groups". The atoms can then be hidden or shown, and the visualization changed, just as for the molecule entries in the Project Tree.
Note that an atom group entry can be renamed. Select the atom group in the Project Tree and invoke the right-click context menu. Here, the Rename option is found.
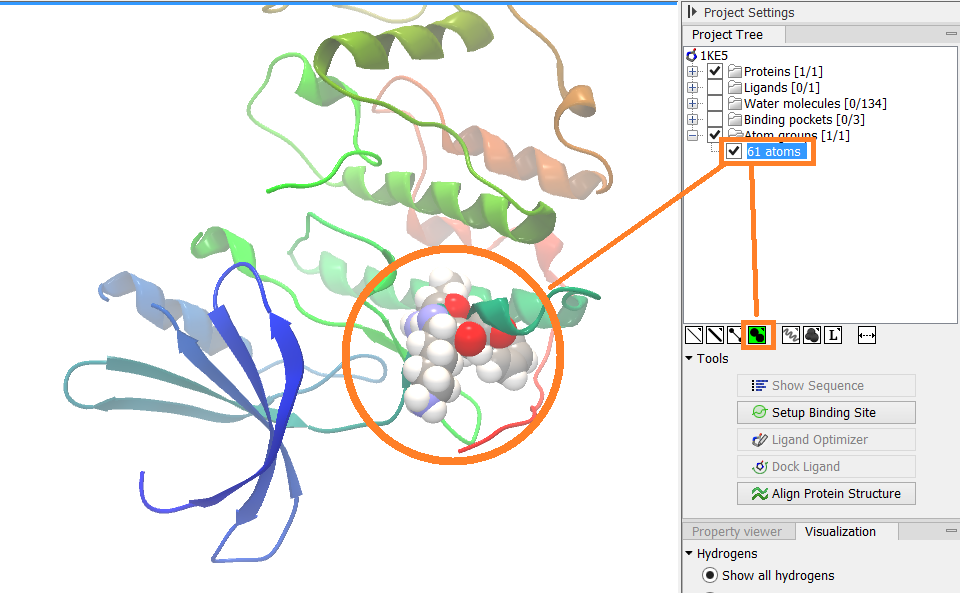
Figure 9.6: An atom group that has been highlighted by adding a unique visualization style.
Create atom group based on atoms selected in 3D view
When atoms are selected in the 3D view, brown spheres indicate which atoms are included in the selection. The selection will appear as the entry "Current" in the Selections category in the Project Tree.
Once a selection has been made, press the "Create Atom Group" button and a context menu will show different options for creating a new atom group based on the selection:
- Selected Atoms. Creates an atom group containing exactly the selected atoms (those indicated by brown spheres). If an entire molecule or residue is selected, this option is not displayed.
- Selected Residue(s)/Molecules. Creates an atom group that includes all atoms in the selected residues (for entries in the protein and nucleic acid categories) and molecules (for the other categories).
- Nearby Atoms. Creates an atom group that contains residues (for the protein and nucleic acid categories) and molecules (for the other categories) within 5 Å of the selected atoms. Only atoms from currently visible Project Tree entries are considered.
- Hydrogen Bonded Atoms. Creates an atom group that contains residues (for the protein and nucleic acid categories) and molecules (for the other categories) that have hydrogen bonds to the selected atoms. Only atoms from currently visible Project Tree entries are considered.
There are several ways to select atoms in the 3D view:
- Double click to select. Click on an atom to select it. When you double click on an atom that belongs to a residue in a protein or in a nucleic acid chain, the entire residue will be selected. For small molecules, the entire molecule will be selected.
- Adding atoms to a selection. Holding down Ctrl while picking atoms, will pile up the atoms in the selection. All atoms in a molecule or category from the Project Tree, can be added to the "Current" selection by choosing "Add to Current Selection" in the context menu. Similarly, entire molecules can be removed from the current selection via the context menu.
- Spherical selection. Hold down the shift-key, click on an atom and drag the mouse away from the atom. Then a sphere centered on the atom will appear, and all atoms inside the sphere, visualized with one of the all-atom representations will be selected. The status bar (lower right corner) will show the radius of the sphere.
- Show Sequence. Another option is to select protein or nucleic acid entries in the Project Tree, and click the "Show Sequence" button found below the Project Tree (Show sequence associated with molecule). A split-view will appear with a sequence editor for each of the sequence data types (Protein, DNA, RNA) (figure 9.7). If you then select residues in the sequence view, the backbone atoms of the selected residues will show up as the "Current" selection in the 3D view and the Project Tree view. Notice that the link between the 3D view and the sequence editor is lost if either window is closed, or if the sequence is modified.
- Align to Existing Sequence. If a single protein chain is selected in the Project Tree, the "Align to Existing Sequence" button can be clicked (Link sequence or sequence alignment to structure). This links the protein sequence with a sequence or sequence alignment found in the Navigation Area. A split-view appears with a sequence alignment where the sequence of the selected protein chain is linked to the 3D structure, and atoms can be selected in the 3D view, just as for the "Show Sequence" option.
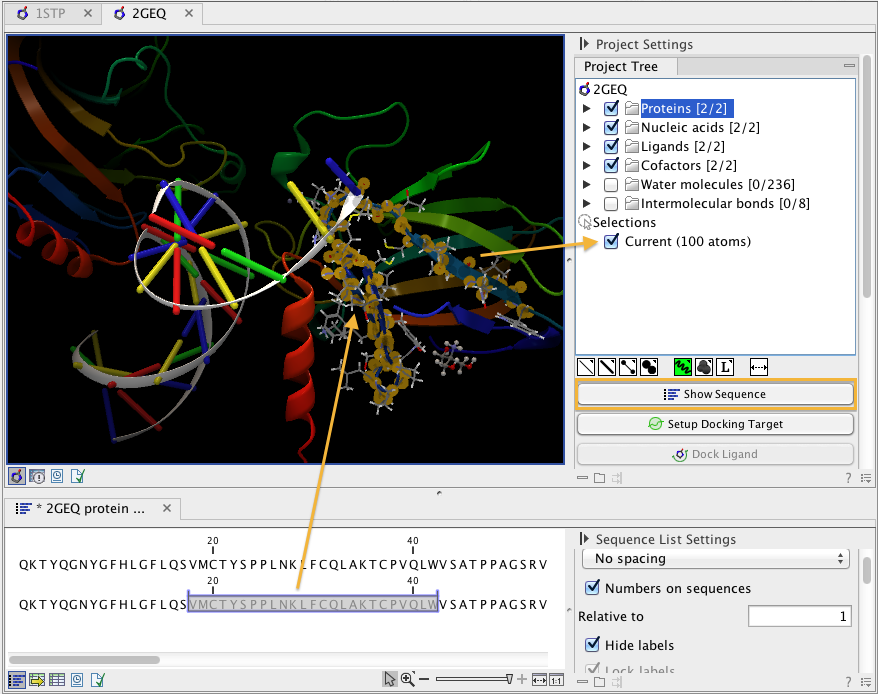
Figure 9.7: The protein sequence in the split view is linked with the protein structure. This means that when a part of the protein sequence is selected, the same region in the protein structure will be selected.
Create atom group based on entries selected in the Project Tree
Select one or more entries in the Project Tree, and press the "Create Atom Group" button, then a context menu will show different options for creating a new atom group based on the selected entries:
- Nearby Atoms. Creates an atom group that contains residues (for the protein and nucleic acid categories) and molecules (for the other categories) within 5 Å of the selected entries. Only atoms from currently visible Project Tree entries are considered. This option is also available on binding pocket entries (binding pockets can only be created in CLC Drug Discovery Workbench).
- Hydrogen Bonded Atoms. Creates at atom group that contains residues (for the protein and nucleic acid categories) and molecules (for the other categories) that have hydrogen bonds to the selected entries. Only atoms from currently visible Project Tree entries are considered.
If a Binding Site Setup is present in the Project Tree (A Binding Site Setup can only be created using CLC Drug Discovery Workbench), and entries from the Ligands or Docking results categories are selected, two extra options are available under the header Create Atom Group (Binding Site). For these options, atom groups are created considering all molecules included in the Binding Site Setup, and thus not taking into account which Project Tree entries are currently visible.
