Running tools and workflows
When CLC Single Cell Analysis Module is installed, the toolbox is populated with Single Cell Analysis specific tools and workflows. These include single cell related importers described in Data import and single cell related exporters described in Data export. In addition, the Single Cell Analysis folder will appear in the toolbox as shown in figure 1.1. To launch a tool or workflow either double-click it in the toolbox or use the quick launch browser (figure 1.10). If you are connected to a CLC Server via your Workbench, you will be asked where you would like to run the analysis. We recommend that you run the analysis on a CLC Server when possible.
Figure 1.10: The Launch button is situated in the top left corner of the workbench.
Running tools
All tools require an input. In many cases this will be an imported Expression Matrix, or, when starting from FASTQ, imported reads.
Clicking the Import button in the top toolbar will bring up a list of supported data types, see figure 1.11. Select the appropriate importer and follow the wizard steps. For additional guidance on import settings please refer to the relevant section of Data import. Save the imported data in the navigation area. This is most easily done by choosing to save and selecting a save location. After import, the sample can be found in the navigation area, ready for analysis.
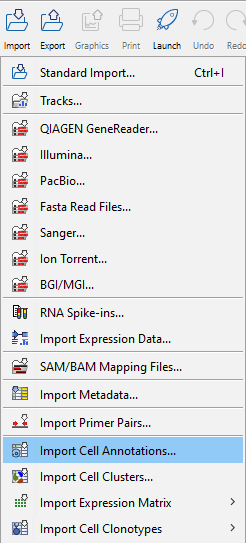
Figure 1.11: Import options with single cell related importers in the lower part. Import Expression Matrix can be expanded to select among the following formats: Cell Ranger HDF5, Loom, MEX, and CSV/TXT. Import Cell Clonotypes can also be expanded to select between the Cell Ranger contig and AIRR formats.
Tools will only run correctly when meaningful input and parameters are provided. If a warning is shown, read it carefully as it will in most cases guide you to the problem.
Select the input based on the description provided in the tool. Although most tools will only allow selection of the correct data type as input, it may still be possible to provide input that does not conform to the tool's expectations. If this happens, the tool will give a warning or gray-out options that cannot be used. The default settings for each tool are suitable for most use cases, but some settings require knowledge of sample preparation or sequencing technology. Most problems arise from inappropriate settings. If a tool produces a report, make sure to inspect it, as it may provide quality control or guidance for how to change settings.
For a full description of running tools, handling results and batch options see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Running_tools_handling_results_batching.html.
Running workflows
CLC Single Cell Analysis Module provides prebuilt workflows for analyzing scRNA-seq data in multiple steps. The workflows can be executed from the toolbox or via the quick launch tool. Input can either be selected from the navigation area or imported using Select files for import. Make sure to select the correct import type. Note: when importing FASTQ, metadata is required to match up read pairs.
Follow the instructions in the wizard steps and save the data by specifying a location in the navigation area. Read more about the provided workflows and how to select proper parameters in section Single cell workflows
For a full description of how to launch workflows and use metadata see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Launching_workflows_individually_in_batches.html.
