When is batch correction appropriate?
If in doubt, apply batch correction only when normalization alone proves unsuitable. The suitability of normalization can often be evaluated by looking at how well cells from different samples are mixed within clusters in a Dimensionality Reduction Plot. Two examples follow:
Example 1: figure 5.17 shows clusters colored by sample, where each sample consists of a single cell type. After batch correction using "Each sample is a batch", the cell types are mixed. This is obviously undesired, so batch correction is inappropriate in this case - any effect of batch on expression is confounded with the effect of cell type on expression, and it is not possible to remove one without also removing the other.
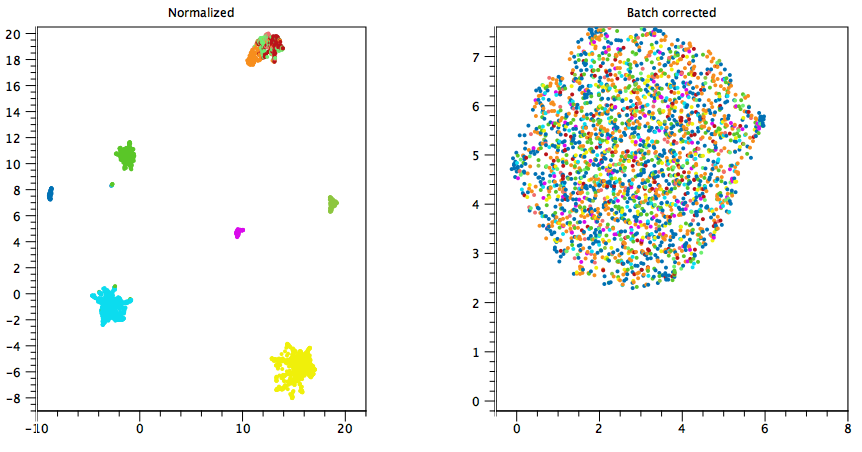
Figure 5.17: An example of when batch correction is undesirable. Each color corresponds to a sample of one cell type. Batch correcting the different samples also removes differences due to cell type, leading to a single cluster.
Example 2: figure 5.18 shows clusters colored by sample. After batch correction using "Each sample is a batch", the clusters are mixed. If the samples described the same tissue type and experimental treatment then we would suspect a batch effect was present, and that batch correction is appropriate. If instead the samples described a difference in experimental treatment, then it would not be possible to determine whether the clusters were separated due to the effect of the treatment, or due to a batch, and deciding whether to apply batch correction would be more difficult.
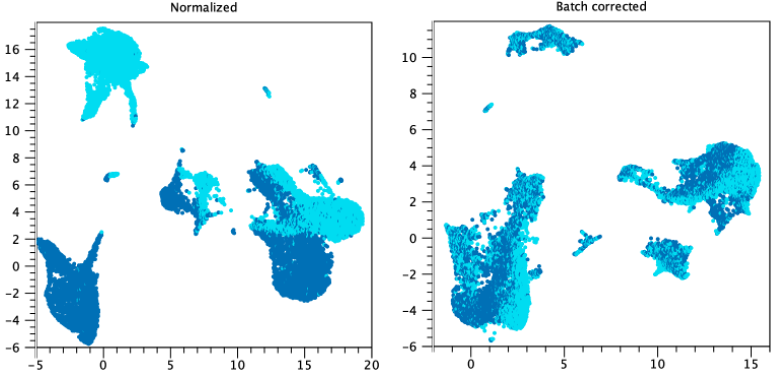
Figure 5.18: An example of when batch correction may be desired. Several clusters can be seen for each of two samples. After batch correction clusters contain a mixture of both samples. Data is from a Seurat tutorial data set (https://satijalab.org/seurat/archive/v3.2/immune_alignment.html).
