Configuring the batch units
When there is only FASTQ file per sample, metadata is not necessary for workflow execution. Typically though, the data is paired and there might be more than one lane per sample. Let us consider the FASTQ files shown in figure 11.1 and the metadata shown in figure 11.2.

Figure 11.1: Example of ten FASTQ files for paired reads, originating from multiple lanes and three libraries.
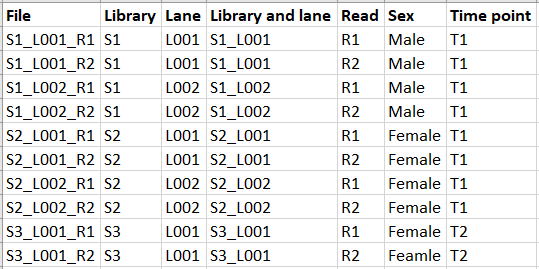
Figure 11.2: Example of metadata for the files from figure 11.1.
The FASTQ files and metadata (in Excel or txt format) can be automatically imported during workflow execution. Choose "Select files for import", select the Illumina importer and enable "Paired reads". For this example, the "Library" metadata column should be used for defining the batch units, as shown in figure 11.3.
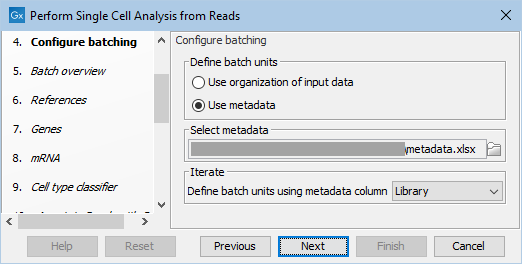
Figure 11.3: Configuring the workflow execution using metadata.
The workflow will automatically associate the input files with the correct rows in the metadata based on the first column and the batch overview will show how the input files are grouped together into batch units (see figure 11.4). The additional metadata columns will be converted to cell annotations.
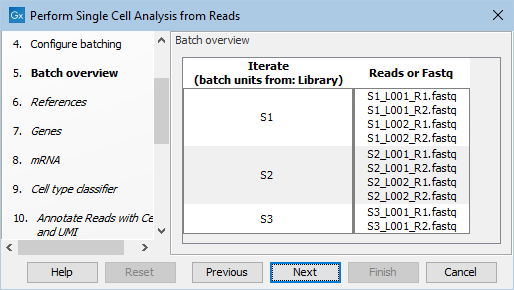
Figure 11.4: Batch overview when importing the FASTQ and metadata files and defining batch units as shown in figure 11.3.
Note that in this workflow it is not possible to freely choose the batch units. Instead, each batch must correspond to one sample. The reason for the restriction is that each read is linked to a cell by the cell barcode. Batching by sample is required in order to inform the workflow that if the same cell barcode is present in multiple files, it is because it is the same cell.
Failing to batch by sample will likely lead to misleading results. For example, in figure 11.3 it would be necessary to batch by library. If we batched by "Time point", then two cells with the same barcode at time point T1 would be treated as being identical, even if one came from sample S1 and the other from sample S2. If, on the other hand, we batched by "Library and Lane", then a cell from sample S1 that was sequenced on both lanes would be split up into two cells - one for each lane.
The workflow combines all inputs to produce just one matrix. All metadata, including "Sex" and "Time point" in the provided example, will be available in the output cell annotations.
The FASTQ and metadata files can also be imported manually and used for the workflow execution.
To avoid the need for metadata, the FASTQ files can be imported manually using the Illumina importer and enabling the "Paired reads" and "Join reads from different lanes". This will result in three sequence lists, one for each library. The workflow can then be configured to "Use organization of input data".
Note that if any of the samples has more than 1 billion paired reads, the "Join reads from different lanes" option should not be used, and metadata is needed for the correct workflow execution.
