Empty droplets filter
In droplet-based data, barcodes can correspond to droplets containing one cell, more cells or no cells at all. In the first dialog of QC for Single Cell, the Empty droplets filter can be enabled and customized to remove the droplets that are detected as being empty. This filter should be skipped for single-cell protocols that are not droplet-based.
Non-zero counts in empty droplets are obtained from ambient (i.e., extracellular) RNA, that can be captured and sequenced during the protocol. Sequenced empty droplets contain significantly fewer reads, and this can be seen as a sharp transition in the rank plot, shown in figure 5.5.
To identify empty droplets, the ambient RNA profile is first modeled from the droplets containing so few reads that they are very likely to be empty. Barcodes are tested against this ambient RNA profile and are assigned simulation-based FDR-corrected p-values, from which non-empty droplets are identified. To reduce the number of tests performed, barcodes with a high number of reads are automatically retained as cell-containing droplets. The threshold for this is usually obtained from the automatically inferred knee from the rank plot (see figure 5.5 and The cell calling algorithm for details).
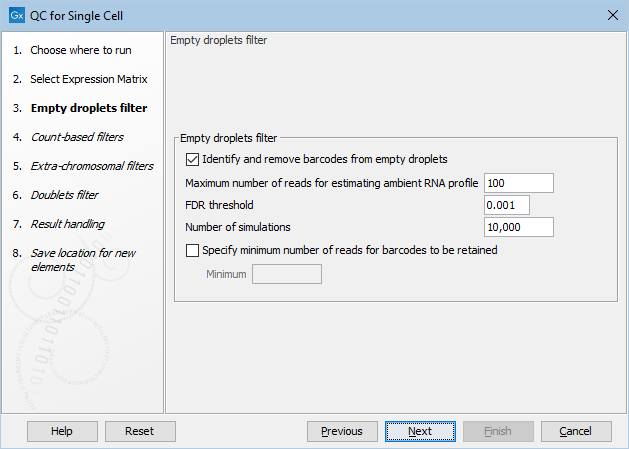
Figure 5.1: The default settings in the Empty droplets filter dialog.
The following options can be adjusted in the Empty droplets filter dialog (figure 5.1):
- Identify and remove barcodes from empty droplets. Enables filtering of the empty droplets. This should be disabled for single-cell protocols that are not droplet-based.
- Maximum number of reads for estimating ambient RNA profile. Barcodes with at most this many total number of reads are considered empty droplets and are used for modeling the ambient RNA.
- FDR threshold. Barcodes with FDR-corrected p-values larger than this are removed as empty droplets.
- Number of simulations. The number of simulations performed for estimating p-values.
- Specify minimum number of reads for barcodes to be retained. Barcodes with a total number of reads above the knee are retained as cell-containing droplets. Enabling this option allows specifying a manual threshold to be used instead of the knee.
- Minimum. The minimum number of reads to be used instead of the knee.
After applying the Empty droplets filter, only barcodes that are identified as non-empty are retained for the remaining filters (see Count-based and extra-chromosomal filters). Note that this filter does not concern the quality of the retained cells. The Empty droplets filter already removes cells with low number of reads, or, by association, low number of expressed features, and enabling the Count-based filters is not strictly necessary. The Extra-chromosomal filters provide the most additional benefit in this situation.
|
Note: It is possible to set options so as to use a simpler Empty droplets filter where only barcodes with a certain number of reads are retained. For example, to retain cells with at least 1000 reads, set Maximum number of reads for estimating ambient RNA profile to 999, enable Specify minimum number of reads for barcodes to be retained and set Minimum to 1000. Note that this has consequences for the number of estimated doublets, see Doublets filter for details. |
For removing droplets containing more than one cell, the Doublets filter can be used, see Doublets filter.
