Setting the sample name
The `demultiplexed reads' are annotated with the sample name, cell barcode and UMI (see Interpreting the output of Annotate Reads with Cell and UMI), all of which are used in downstream tools. When jointly analyzing scTCR-seq with matched scRNA-seq data, it is important that reads originating from the same sample are annotated with the same sample name. This can be configured using the `Sample name' option.
When the tool is run from the Toolbox, the `Sample name' option supports the placeholder {nameOrBatchId}, which sets the sample name to the name of the input. When the tool is run as part of a workflow, this option will set the sample name to the name of the input for simple workflows, and to the name of the batch identifier for batching/iterating workflows. Additional placeholders are available when the tool is run as part of a workflow (figure 4.4).
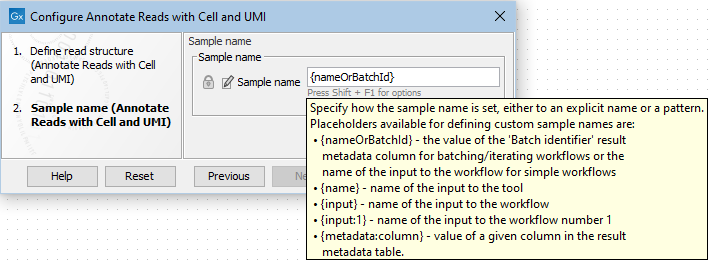
Figure 4.4: Options for configuring the `Sample name' when the Annotate Reads with Cell and UMI is run in a workflow.
Note that `Sample name' can also be set to an explicit name, such as `Individual_1', or to a complex pattern such as `Individual_{nameOrBatchId}_1'.
