Configuring the batch units with multiple input types
The Immune Repertoire and Expression Analysis from Reads (10xVDJ) workflow requires metadata containing information about the sample of origin and data type: scRNA-seq or scTCR-seq (figure 11.8). This can be created a priori, or it can imported directly from an Excel or txt file during workflow execution. For more information about Metadata Tables, see
https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manuall=Metadata.html.
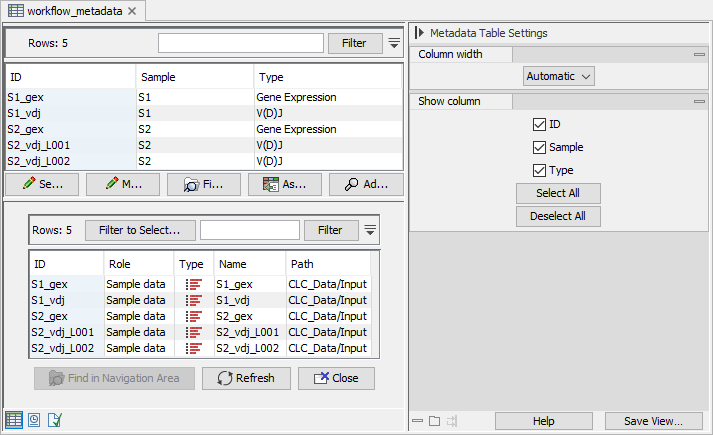
Figure 11.8: A Metadata Table associating the reads to samples and data type. The sample of origin is given in the "Sample" column, while the data type is given in the "Type" column, with values "Gene Expression" for scRNA-seq data, and "V(D)J" for scTCR-seq data. The associated elements are opened on the bottom.
During the workflow execution, it must be configured which metadata columns define the sample: "Iterate over Sample" and data type: "Iterate over RNA and TCR", and which values from the data type column correspond to scRNA-seq and scTCR-seq data, respectively. For more details on configuring workflow execution with metadata, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Batching_part_workflow.html. Make sure to inspect the batch overview to check that the analysis will be performed correctly.
Figures 11.9 and 11.10 show how the workflow should be configured using the input reads and metadata from figure 11.8, and the resulting batch overview is given in figure 11.11.
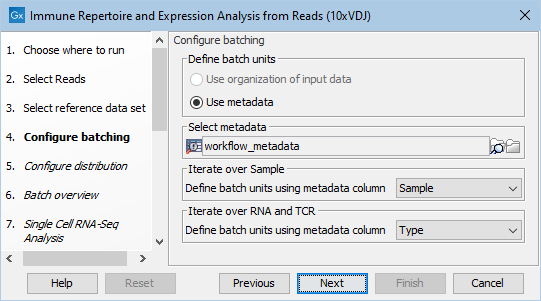
Figure 11.9: Configuring sample and data type during workflow execution using the metadata from figure 11.8.
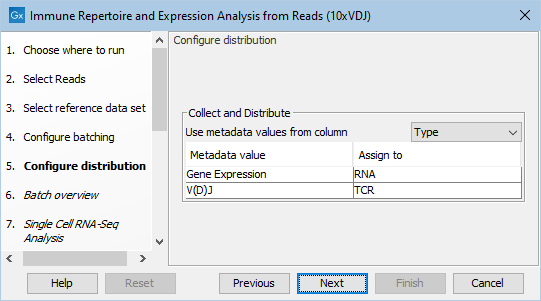
Figure 11.10: Configuring scRNA-seq and scTCR-seq reads distribution during workflow execution using the metadata from figure 11.8.
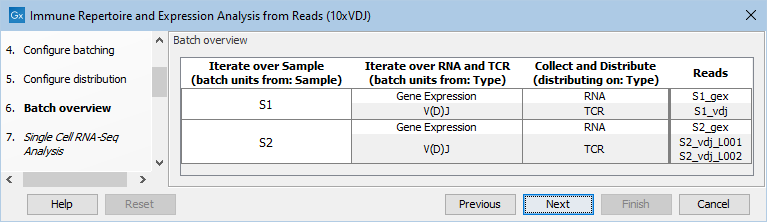
Figure 11.11: Batch overview for the configuration shown in figures 11.9 and 11.10.
An additional example for importing the reads and metadata during workflow execution can be found in Configuring the batch units.
