Import Cell Clonotypes
The following cell clonotypes formats produced by Cell Ranger can be imported into a Cell Clonotypes (- Import Cell Clonotypes in Cell Ranger Contig Format (
 ): CSV contig format;
): CSV contig format;
- Import Cell Clonotypes in AIRR Format (
 ): AIRR format.
): AIRR format.
The importers can be found here:
Import (![]() ) | Import Cell Clonotypes (
) | Import Cell Clonotypes (![]() )
)
Most options are common to both importers. The following options can be adjusted (figure 2.6):
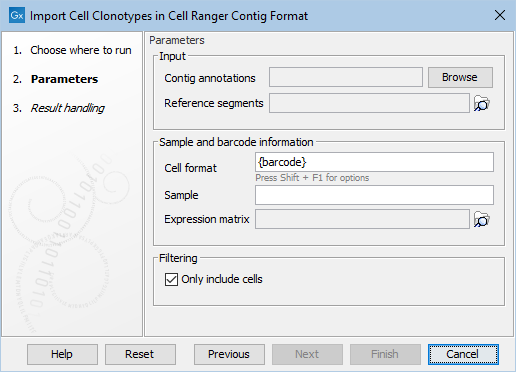
Figure 2.6: Options in the dialog of Import Cell Clonotypes in Cell Ranger Contig Format. The same options are present in the dialog of Import Cell Clonotypes in AIRR Format, where "Contig annotations" is named "AIRR rearrangements".
- Contig annotations / AIRR rearrangements. The input .csv or .tsv file, respectively, following the official Cell Ranger format.
- Reference segments (Optional). A reference data element downloaded from the Reference Data Manager (see The Reference Data Manager) containing the corresponding QIAGEN V and J genes. When supplied, the gene names in the input file will be mapped to those used in the provided element. This is important when comparing imported Cell Clonotypes with those produced by the CLC Single Cell Analysis Module, see Compare Single Cell Immune Repertoires.
- Cell format. How to extract the barcode, and optionally the sample name, from the column containing the barcode. For example:
- If the cell format is set to "{barcode}-1", then "AAGCT-1" will be interpreted as describing a cell with barcode "AAGCT".
- If the cell format is set to "{sample}-{barcode}", then "demo-AAGCT" will be interpreted as describing a cell from sample "demo" with barcode "AAGCT".
- Sample (Optional). Sets a custom sample name.
- Expression matrix (Optional). The sample name will be obtained from the supplied Expression Matrix. The importer does not check that the barcodes present in the input file match those in the matrix. If the Expression Matrix contains multiple samples, the importer will fail with a relevant message.
If none of the above three options provides a sample name, the importer sets it to the name of the input file.
The importer does not allow setting the sample name using more than one of these three options.
It is important to set an appropriate sample name when matched scRNA-seq data is available, see Convert Clonotypes to Cell Annotations.
- Only include cells. When this is enabled, barcodes that were not called as cells are not imported.
