Import Cell Clusters
The Import Cell Clusters tool can import clusters for each cell. The importer produces Cell Clusters (Often the same file can be imported as either Cell Clusters or Cell Annotations. The principal advantage of Cell Clusters is that they can be edited within the Dimensionality Reduction Plot.
The importer can be found here:
Import (![]() ) | Import Cell Clusters (
) | Import Cell Clusters (![]() ).
).
The following options are available:
- Data file. A single file in .csv or .xlsx format. The first row in the file is used as the name of the columns of the imported table. Each subsequent row in the file should describe a cell (though empty lines are ignored). Cells are identified by a combination of their barcode e.g. "AAGCT" and their sample name.
- First column defines sample. When this is enabled, the first column in the file will be treated as the sample name, and the second column defines the barcode. Subsequent columns are annotations on the cells.
- Cell format. How to extract the barcode, and optionally also the sample name, from the column containing the barcode. For example:
- If the cell format is set to "{barcode}-1" then a row that contains "AAGCT-1" in this column will be interpreted as describing a cell with barcode "AAGCT".
- If the cell format is set to "{sample}-{barcode}" then a row that contains "demo-AAGCT" in this column will be interpreted as describing a cell from sample "demo" with barcode "AAGCT".
- Expression matrix. This is optional when First column defines sample is enabled, or when Cell format describes how to extract the sample name. When an Expression Matrix is supplied:
- The sample name is taken from the matrix. If the file provides different sample names than the matrix, then the importer fails with an error. This can be useful when checking that the file being imported matches the supplied matrix.
- Rows in the file describing cells that are not in the matrix are skipped.
- Map clusters to QIAGEN Cell Ontology. When this is enabled, clusters will be translated, if possible, to the QIAGEN Cell Ontology (see The QIAGEN Cell Ontology). The translation attempts to match each cluster with a cell type from the ontology based on the name and known synonyms. For example, `alveolar epithelial cells' are also called `pneumocytes'. If the data file contains an `alveolar epithelial cells' cluster (as in figure 2.4) and this option is selected, then in the imported file the cluster will be named `pneumocytes' (see figure 2.5). This option can be useful when standardizing cell annotations from different sources. It is especially recommended if the imported data will be used to extend a QIAGEN Cell Type Classifier using the Train Cell Type Classifier tool (Train Cell Type Classifier).
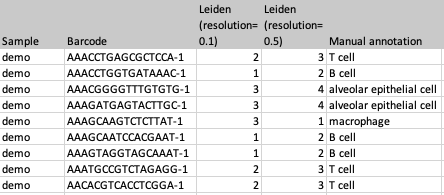
Figure 2.4: An example of a file that could be imported with Import Cell Clusters. The file contains three different clusterings.

Figure 2.5: The result of importing the file shown in figure 2.4 using the option `Map clusters to QIAGEN Cell Ontology'. Note that all cell types have been translated to terms in the QIAGEN Cell Ontology. For example, `T cells' have been standardized to `T lymphocytes', and `alveolar epithelial cells' have been standardized to `pneumocytes'.
