Doublets filter
In droplet-based data, barcodes can correspond to droplets containing two or more cells. In this dialog of QC for Single Cell, the Doublets filter can be enabled and customized to remove the droplets that are detected as containing two cells. This filter should be skipped for single-cell protocols that are not droplet-based.
|
Note: The Doublets filter can only be used together with the Empty droplets filter. |
Note that QC for Single Cell cannot remove droplets containing more than two cells. However, these are expected to be present at negligible rates.
There are two types of doublets:
- homotypic doublets are formed by two cells with similar expression profiles;
- heterotypic doublets are formed by two cells with different expression profiles.
Doublet-removal software, which relies on gene expression to detect doublets, cannot identify homotypic doublets, as their expression profiles are indistinguishable from those of other cells. Alternative approaches are required to detect homotypic doublets, such as cell hashing [Stoeckius et al., 2018] and SNPs in multiplexed samples [Kang et al., 2018].
The Doublets filter simulates heterotypic doublets by averaging the expression of two random barcodes that are sufficiently different from each other. These artifical doublets are then used for predicting which of the input barcodes are doublets.
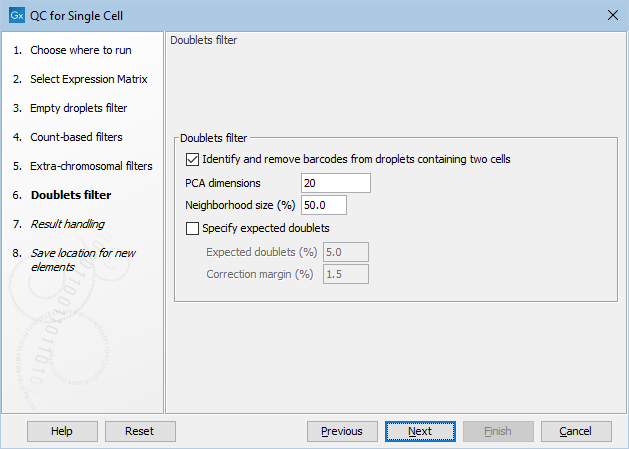
Figure 5.4: The default settings in the Doublets filter dialog.
The following options can be adjusted in the Doublets filter dialog (figure 5.4):
- Identify and remove barcodes from droplets containing two cells. Enables filtering of the doublets. This should be disabled for single-cell protocols that are not droplet-based.
- PCA dimensions. The number of PC dimensions to be used when reducing the dimensions of the expression data.
- Neighborhood size (%). Simulated doublets are obtained from barcodes that are not in each other's neighborhood. The size of the neighborhood is specified as % of input barcodes. Note that this is relative to the number of barcodes that pass all previous filters of QC for Single Cell. The optimal neighborhood size is data-set specific and would typically depend on the number of clusters in the data.
- Specify expected doublets. Enable this option to specify approximately how many doublets are present in the data. This option should be used whenever a reasonable expectation is known, as it is very important for an accurate detection of doublets.
- Expected doublets (%). The percentage of barcodes that are expected to be doublets, relative to the number of captured cells. If 'Specify expected doublets' is disabled, this is set to
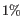 per
per 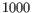 captured cells, which is roughly the doublet rate for 10x data.
captured cells, which is roughly the doublet rate for 10x data.
- Correction margin (%). The percentage of predicted doublets will lie in the interval given by 'Expected doublets (%)'
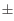 'Correction margin (%)'. If 'Specify expected doublets' is disabled, this is set to half of the value of 'Expected doublets (%)'.
'Correction margin (%)'. If 'Specify expected doublets' is disabled, this is set to half of the value of 'Expected doublets (%)'.
|
Note: Expected doublets (%) is relative to the number of captured cells and not to the number of high quality cells. This is estimated as the number of barcodes passing the Empty droplets filter. The Doublets filter receives as input only the high quality cells that also pass the Count-based filters and Extra-chromosomal filters. |
For more details on how doublets are detected, see The doublet calling algorithm.
