Locus table
The locus table (figure 13.7) displays all genotype track loci, one per row. All available genotype track annotations refer to either the locus-, variant- or haplotype-level. This is indicated in the side panel by the superscript: L = Locus annotation, V = Variant annotation, and H = Haplotype annotation. The displayed columns in the locus table are locus annotations by default. Loci where filters apply are displayed when the option 'Show non-PASS' is enabled. Blue columns are sample specific annotations, corresponding to FORMAT annotations in VCF (for a more detailed description of VCF compatibility, see section 13.2.7).
Below are descriptions of general locus table annotations. In addition to the general locus table annotations, the genotype track also contains some basic variant annotations. Read about them here: https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Variant_tracks.html.
- Chromosome
- The name of the reference sequence where the variant locus is situated.
- Region
- The reference sequence positions of the variant locus. The region may be either a 'single position', a 'region' or a 'between position region'.
- Reference
- The reference allele nucleotide sequence of the locus. Maximally 20 nucleotides are shown. Longer sequences can be obtained in their entirety by copy-pasting the table cell.
- Alt allele
- Alternatives to the reference allele.
- Filter
- List of filters that are directly or indirectly applicable to the locus. The value 'PASS' specifies that the locus passed all filters.
- Alt length
- Length of the part of the allele that is altered relative to the reference, e.g. for deletions the number of deleted symbols and for insertions the number of inserted symbols. Leading and trailing sequence identical to the reference is not included.
- Length
- Total allele length within this locus. Includes any leading and trailing sequence identical to the reference.
- Alt count
- Number of non-reference allele variants at this locus.
- Variant filter
- Allele variant specific filters.
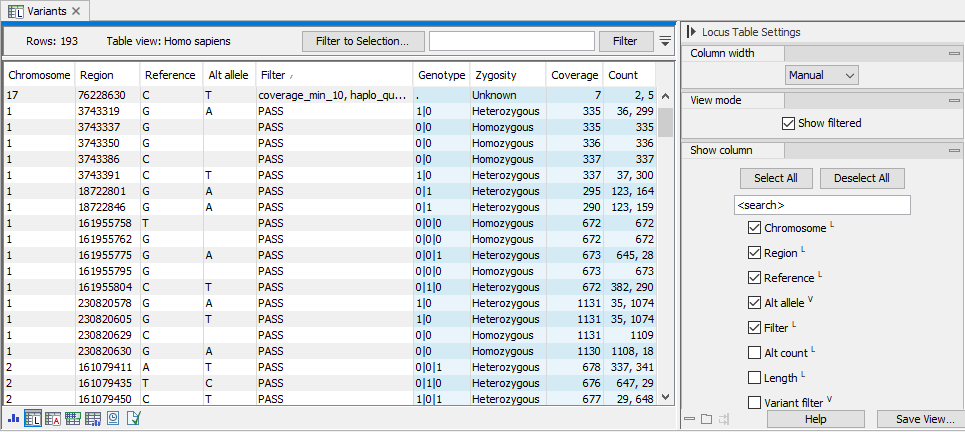
Figure 13.7: The locus table. To go to the locus table view of a genotype track as shown here, select the table icon with an L in the lower left corner. Note that in this example, "Show non-PASS" has been checked in the side panel and filtered loci are shown in the table. Columns displayed can be adjusted by selecting and deselecting columns in the side panel to the right.
Sample annotations
The following annotations are available in all sample genotype tracks.
- Genotype
- The VCF genotype that lists alleles present in the sample genome at this locus. Alleles with applied filters are hence excluded from the genotype.
The genotype is encoded as allele values separated by either of '/' or '|'. The allele values are 0 for the reference allele, 1 for the first allele listed in the 'Alt allele' annotation, 2 for the second allele listed in the 'Alt allele' annotation and so on (figure 13.7). For diploid calls examples could be 0/1, 1|0, or 1/2, etc.
Genotype phasing compatible with all ploidy levels
One way to interpret VCF phasing encoded in the genotype (GT) field, is to consider the separators (/ or |) as phasing flags for the following allele, similar to the way phasing is encoded in BCF. For example in case of GT=0/1|2/3 we would know that 1 and 3 are unphased while 2 is phased. This interpretation, however, leaves the phasing status of the first GT allele poorly defined.
Assumptions are frequently made about the phasing status of the first allele in diploid scenarios. For example GT=0|1 is commonly interpreted to mean that both alleles are phased, and GT=0/1 that both alleles are unphased.
Considering the above, the genotype is written so that:
- At loci where all alleles except the first have same phasing status: the appropriate phasing flag is prepended. For example GT=|0/1/2 or GT=/0|1|2 or GT=/0|1
- At loci where alleles have mixed phasing status, and the first allele is phased: the appropriate phasing flag is prepended. For example GT=|0|1/2 or GT=|0/2
- In the case of haploid loci: if the allele is phased, either the appropriate phasing flag is prepended (e.g. GT=|1), or a missing allele is appended with the appropriate phasing flag (e.g. GT=1|.).
Thus, when encountering a genotype where the first allele has no prepended phasing flag, we can determine phasing status of the first allele to be:
- phased, if phasing flags are present and indicate that all other alleles are phased.
- unphased, in all other cases.
- Phase set
- Identifier for a set of phased genotypes that together describe a set of overlapping haplotypes.
- Zygosity
- The zygosity of the sample genome locus (figure 13.7). This will be 'Homozygous' when only one allele variant is called at the locus, and 'Heterozygous' when more than one variant is called.
- Number of alleles
- Total number of alleles in called genotype.
- Complex
- Complex region, if locus is part of a complex of overlapping loci.
