Symbolic representation in sequence
view
In the Side Panel
of normal sequence views (![]() ), you will find an extra
group under Nucleotide info called Secondary Structure. This
is used to display a symbolic representation of the secondary
structure along the sequence (see figure 24.19).
), you will find an extra
group under Nucleotide info called Secondary Structure. This
is used to display a symbolic representation of the secondary
structure along the sequence (see figure 24.19).
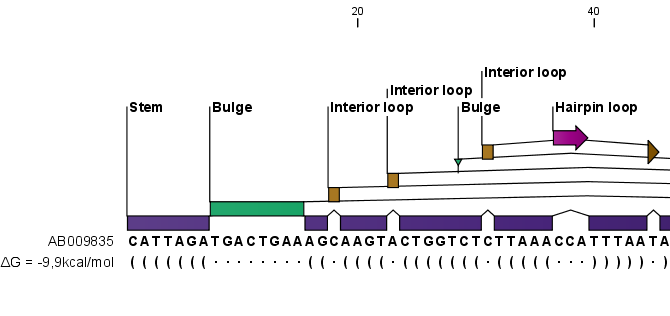
Figure 24.19: The secondary structure visualized below the sequence and with annotations shown above.
The following options can be set:
- Show all structures. If more than one structure is predicted, this option can be used if all the structures should be displayed.
- Show first. If not all structures are shown, this can be used to determine the number of structures to be shown.
- Sort by. When you select to display e.g. four out of
eight structures, this option determines which the "first four"
should be.
- Sort by
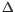 G.
G.
- Sort by name.
- Sort by time of creation.
- Sort by
- Base pair symbol. How a base pair should be represented (see figure 24.19).
- Unpaired symbol. How bases which are not part of a base pair should be represented (see figure 24.19).
- Height. When you zoom out, this option determines the height of the symbols as shown in figure 24.20 (when zoomed in, there is no need for specifying the height).
- Base pair probability. See Probability-based coloring).
When you zoom in and out, the appearance of the symbols change. In figure 24.19, the view is zoomed in. In figure 24.20 you see the same sequence zoomed out to fit the width of the sequence.
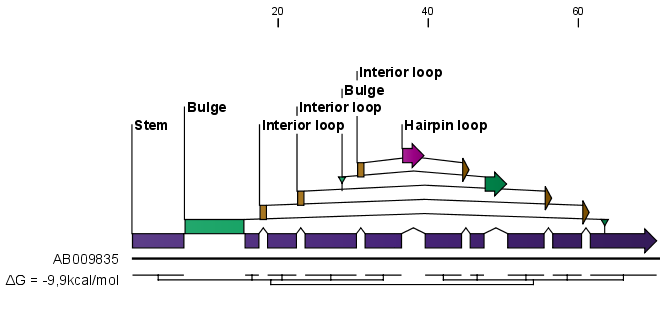
Figure 24.20: The secondary structure visualized below the sequence and with annotations shown above. The view is zoomed out to fit the width of the sequence.
